Constructor Method for the Enriched Heatmap
EnrichedHeatmap.rdConstructor Method for the Enriched Heatmap
EnrichedHeatmap(mat,
col,
top_annotation = HeatmapAnnotation(enriched = anno_enriched()),
row_order = order(enriched_score(mat), decreasing = TRUE),
pos_line = TRUE,
pos_line_gp = gpar(lty = 2),
axis_name = NULL,
axis_name_rot = 0,
axis_name_gp = gpar(fontsize = 10),
border = TRUE,
cluster_rows = FALSE,
row_dend_reorder = -enriched_score(mat),
show_row_dend = FALSE,
show_row_names = FALSE,
heatmap_legend_param = list(),
...)Arguments
- mat
A matrix which is returned by
normalizeToMatrix.- col
Color settings. If the signals are categorical, color should be a vector with category levels as names.
- top_annotation
A special annotation which is always put on top of the enriched heatmap and is constructed by
anno_enriched.- row_order
Row order. Default rows are ordered by enriched scores calculated from
enriched_score.- pos_line
Whether draw vertical lines which represent the positions of
target?- pos_line_gp
Graphic parameters for the position lines.
- axis_name
Names for axis which is below the heatmap. If the targets are single points,
axis_nameis a vector of length three which corresponds to upstream, target itself and downstream. If the targets are regions with width larger than 1,axis_nameshould be a vector of length four which corresponds to upstream, start of targets, end of targets and downstream.- axis_name_rot
Rotation for axis names.
- axis_name_gp
Graphic parameters for axis names.
- border
Whether show the border of the heatmap?
- cluster_rows
Clustering on rows are turned off by default.
- show_row_dend
Whether show dendrograms on rows if hierarchical clustering is applied on rows?
- row_dend_reorder
Weight for reordering the row dendrogram. It is reordered by enriched scores by default.
- show_row_names
Whether show row names?
- heatmap_legend_param
A list of settings for heatmap legends.
atandlabelscan not be set here.- ...
Other arguments passed to
Heatmap.
Details
The enriched heatmap is essentially a normal heatmap but with several special settings. Following parameters are set with pre-defined values:
cluster_columnsenforced to be
FALSEshow_column_namesenforced to be
FALSEbottom_annotationenforced to be
NULL
EnrichedHeatmap calls Heatmap, thus, most of the
arguments in Heatmap are usable in EnrichedHeatmap such as
to apply clustering on rows, or to split rows by a data frame or k-means clustering. Users can also
add more than one heatmaps by + operator. Enriched heatmaps and normal heatmaps can be
concatenated mixed.
For detailed demonstration, please go to the vignette.
Value
A Heatmap-class object.
Examples
load(system.file("extdata", "chr21_test_data.RData", package = "EnrichedHeatmap"))
mat3 = normalizeToMatrix(meth, cgi, value_column = "meth", mean_mode = "absolute",
extend = 5000, w = 50, smooth = TRUE)
#> All signal values are within [0, 1], so we assume it is methylation
#> signal. Automatically set limit [0, 1] to the smoothed values. If this
#> is not the case, set argument `limit = NA` in the function to remove
#> the limits. Set `verbose = FALSE` to turn off this message.
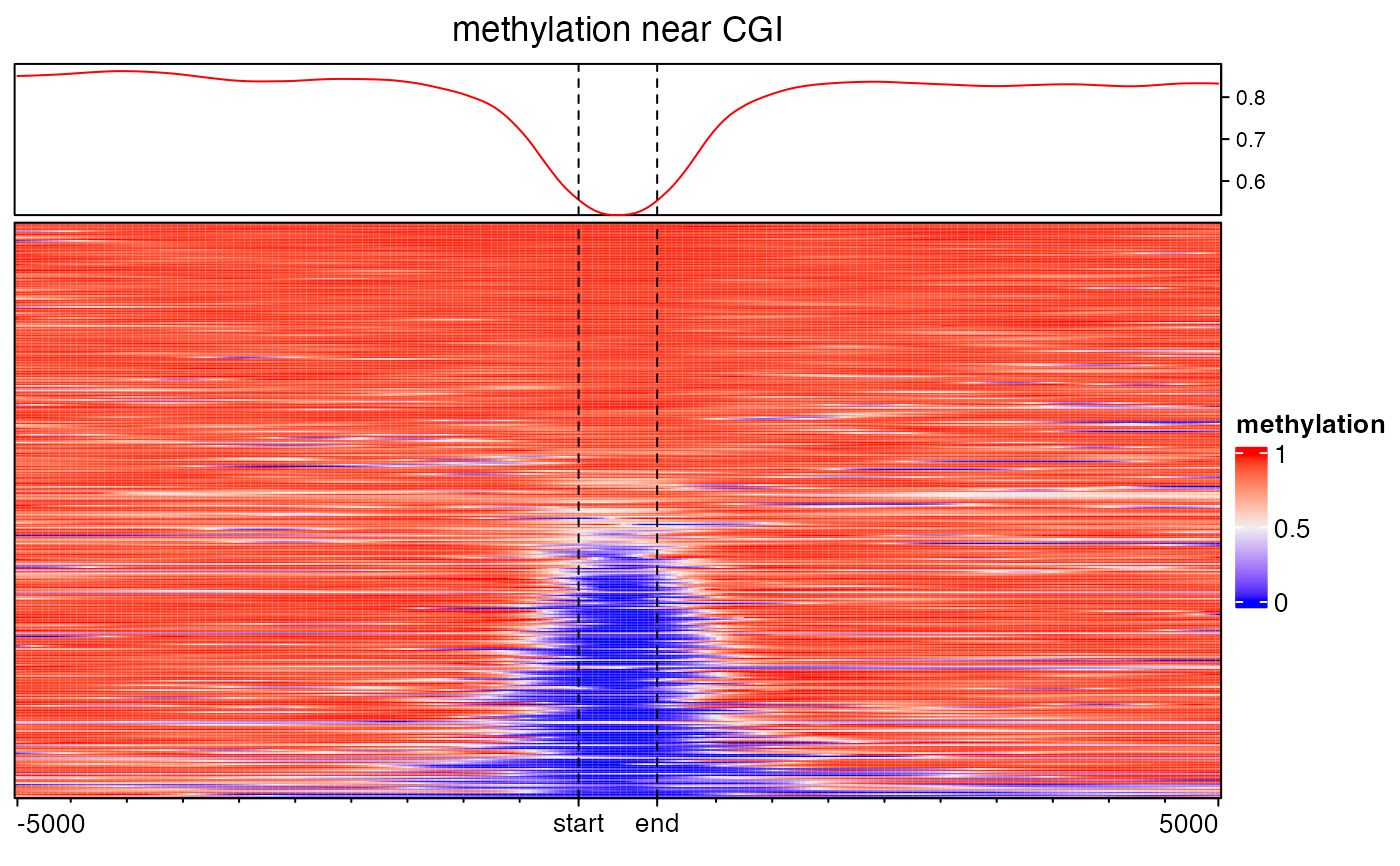
EnrichedHeatmap(mat3, name = "methylation", column_title = "methylation near CGI")
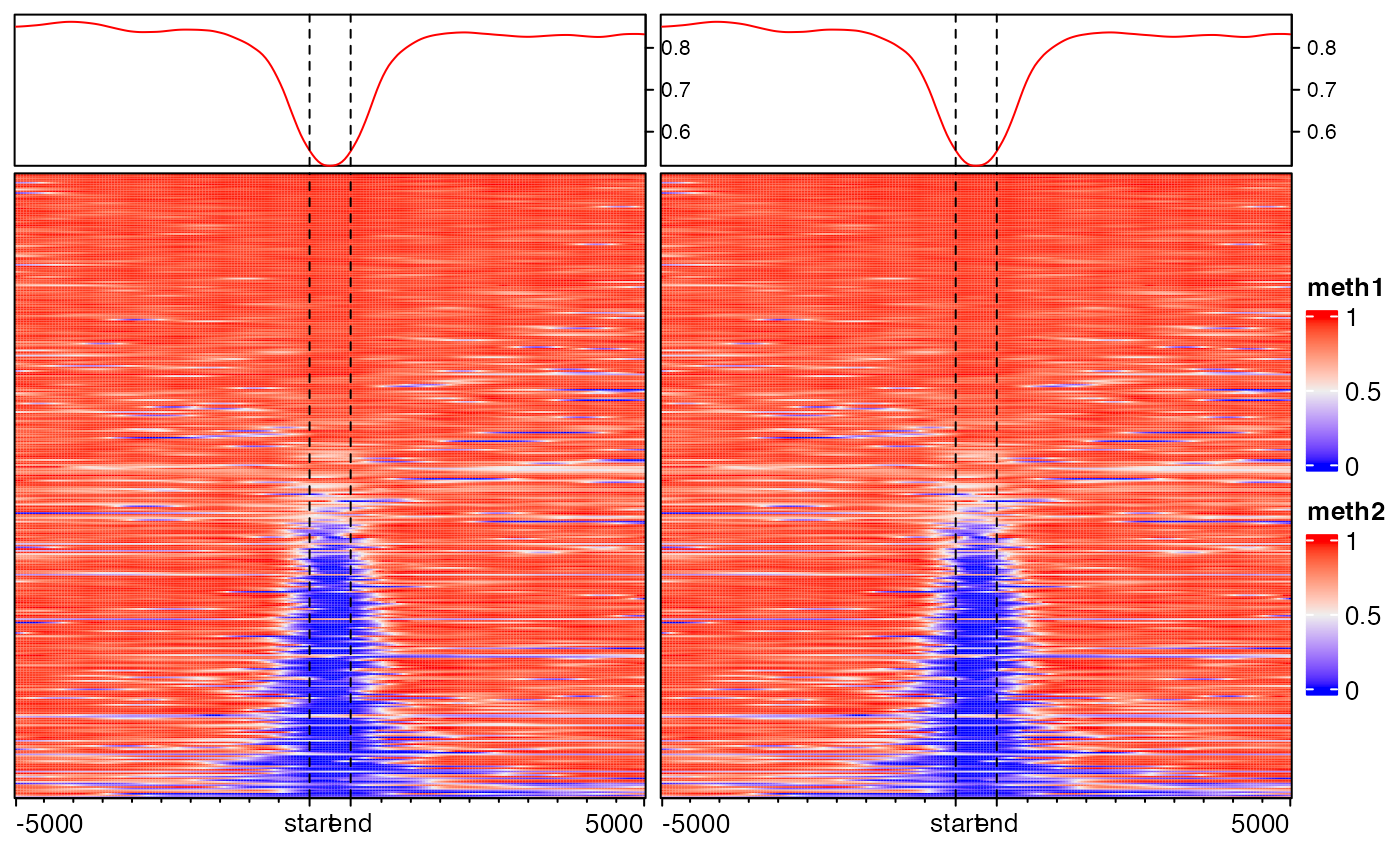
 EnrichedHeatmap(mat3, name = "meth1") + EnrichedHeatmap(mat3, name = "meth2")
EnrichedHeatmap(mat3, name = "meth1") + EnrichedHeatmap(mat3, name = "meth2")
 # for more examples, please go to the vignette
# for more examples, please go to the vignette