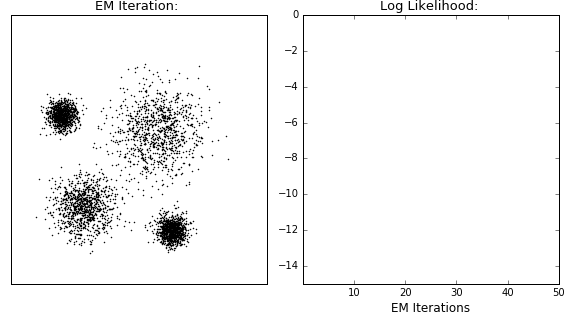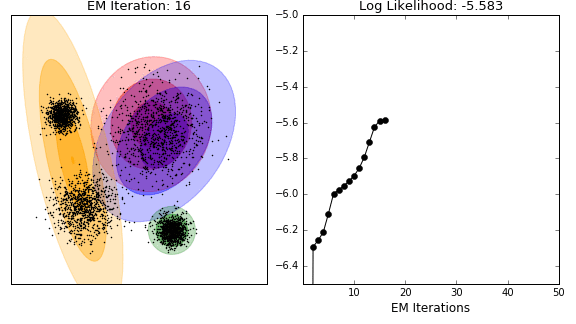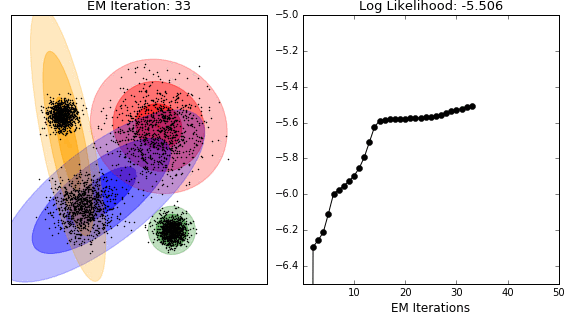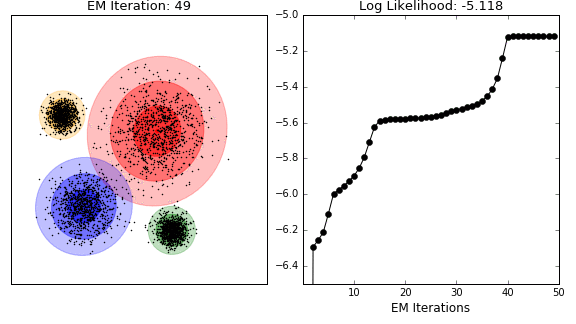
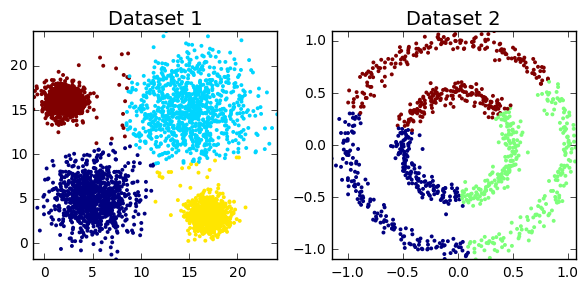
Unsupervised analysis
Statistical analysis without any knowledge for the labeling of samples
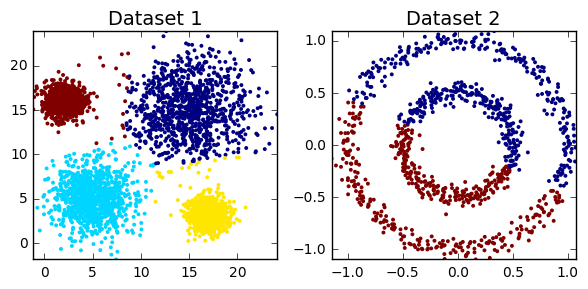
The main aim is to find subgroups
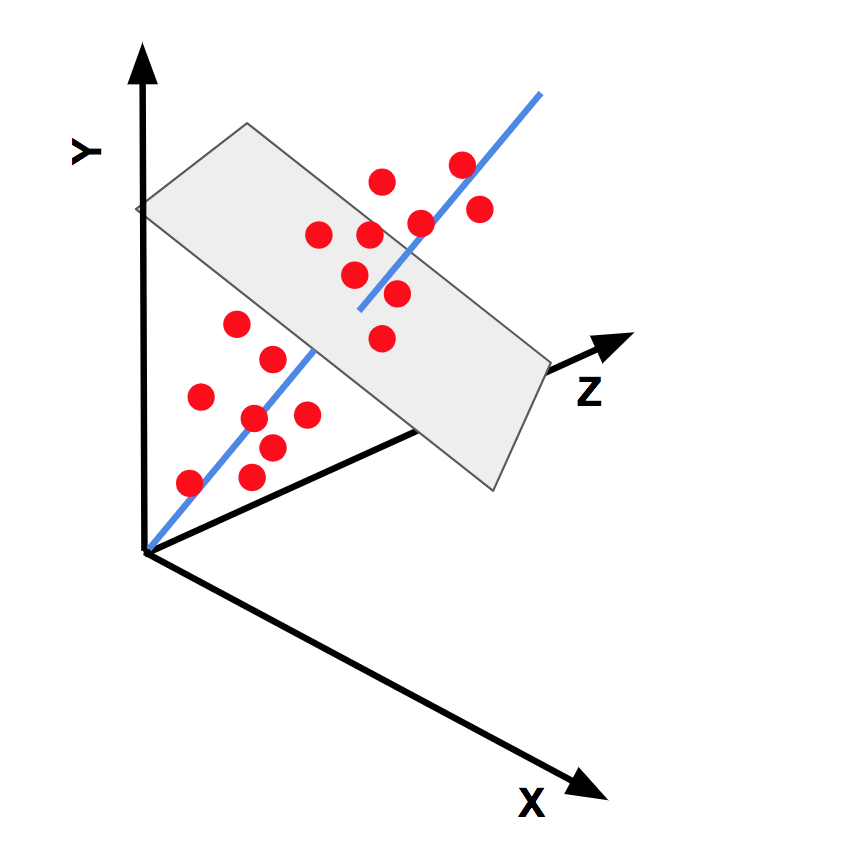
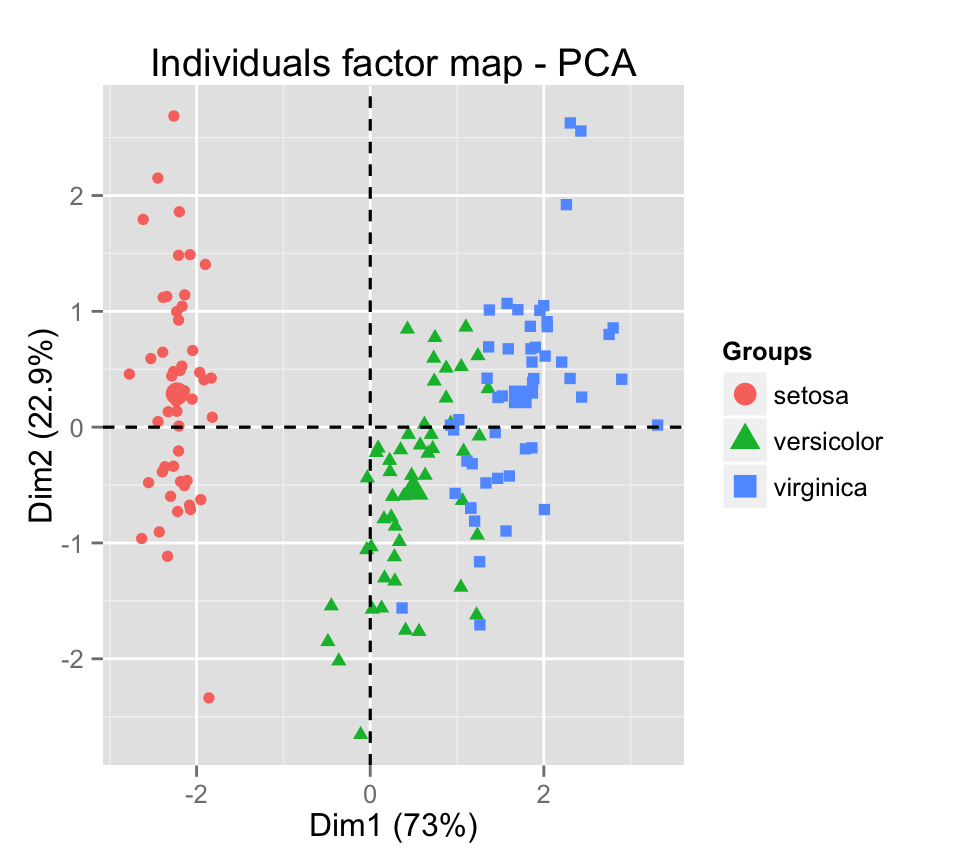
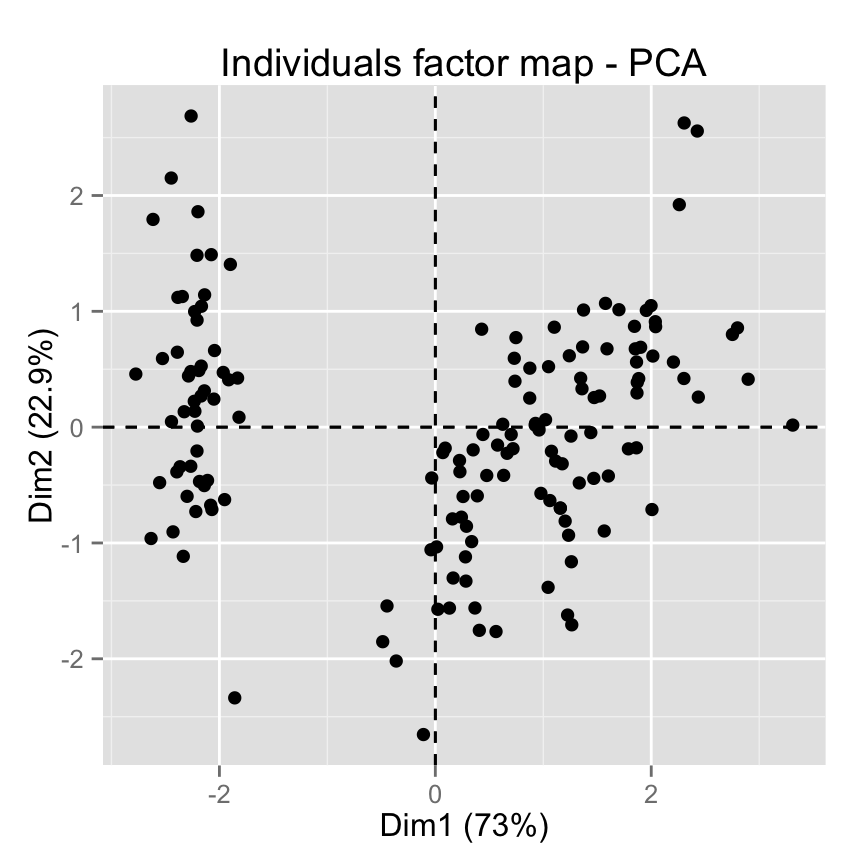
- Dimension reduction
- Principle component analysis (PCA)
- Multiple dimensional scaling (MDS)
- Clustering
- hierarchical clustering
- partitioning (k-means)
- model-based clustering
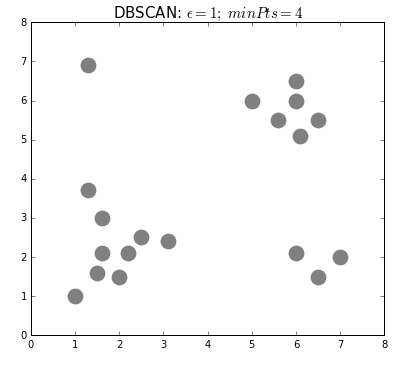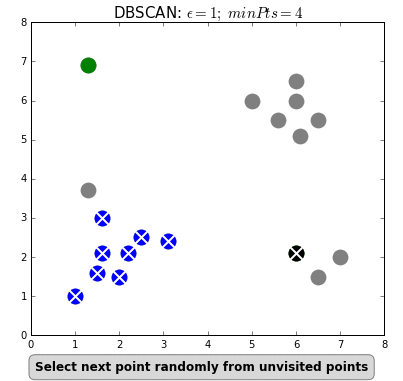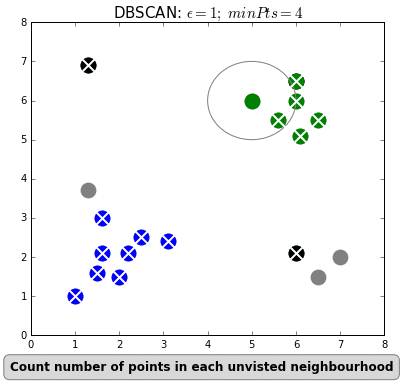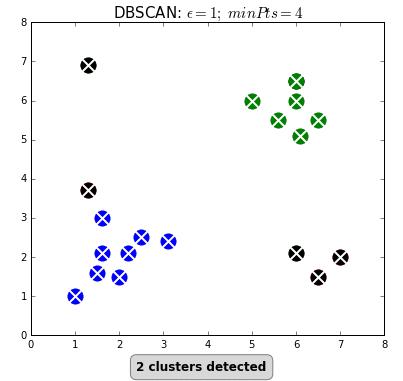
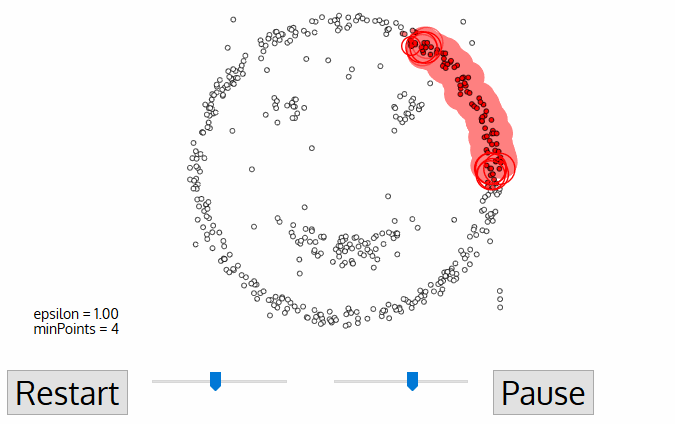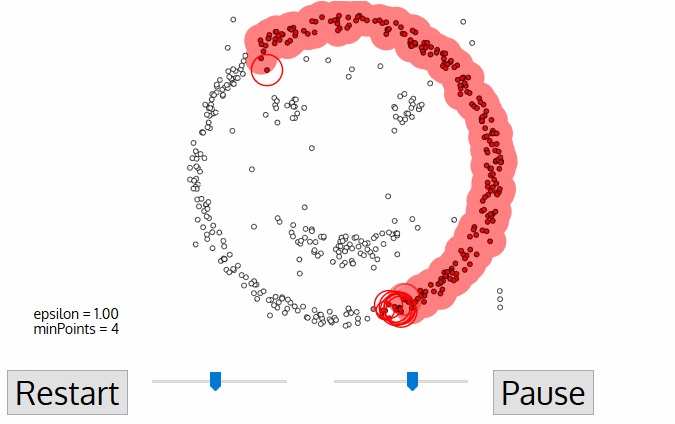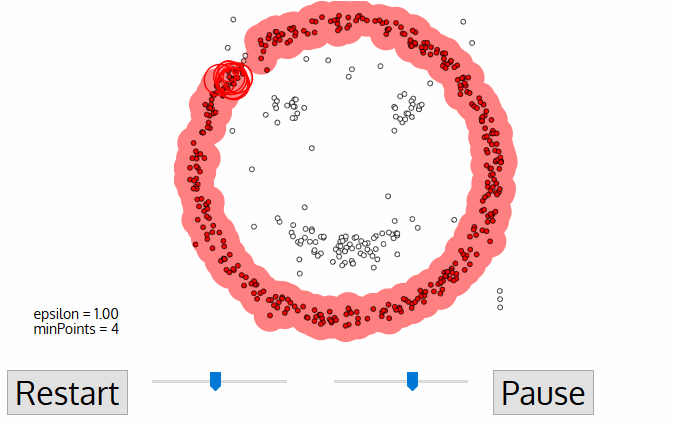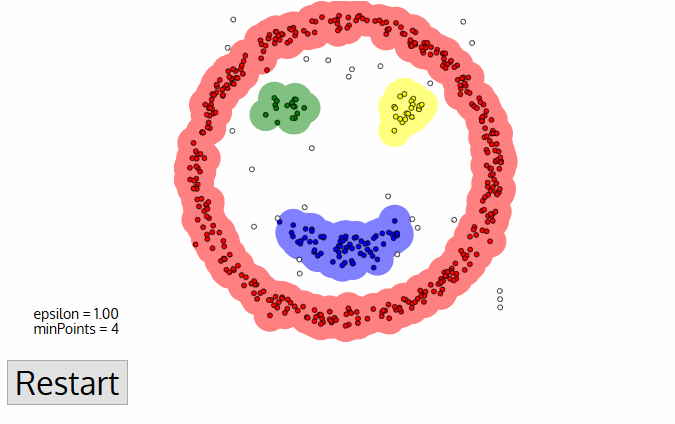
- density-based clustering
- consensus clustering