circlize provides general solutions for circular visualization.
Basic
| 软件包 | 链接 |
|---|---|
| Language | R |
| CRAN | https://CRAN.R-project.org/package=circlize |
| GitHub | https://github.com/jokergoo/circlize |
| Documentation | https://jokergoo.github.io/circlize_book/book/ |
| Publication | Zuguang Gu, et al., circlize Implements and enhances circular visualization in R. Bioinformatics 2014. |
Example
Simple xy-coordination system.
library(circlize)
factors = letters[1:7]
circos.initialize(factors, xlim = c(0, 1))
circos.trackPlotRegion(ylim = c(0, 1), track.height = 0.4, panel.fun = function(x, y) {
circos.points(runif(100), runif(100), cex = 0.5)
})
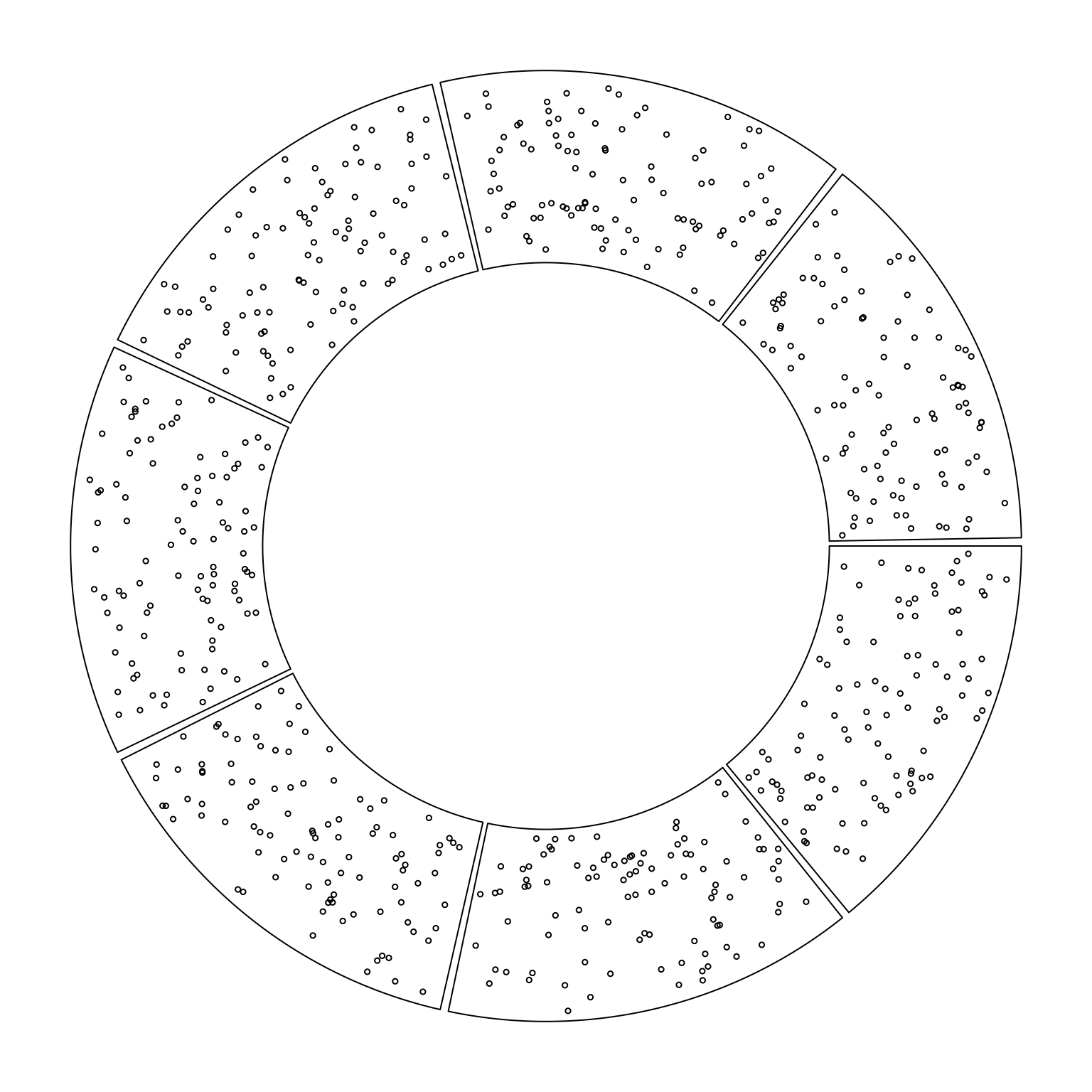
circos.clear()
Genome-level visualization.
load(paste(system.file(package = "circlize"), "/extdata/DMR.RData", sep = ""))
circos.initializeWithIdeogram(plotType = c("axis", "labels"))
bed_list = list(DMR_hyper, DMR_hypo)
circos.genomicRainfall(bed_list, pch = 16, cex = 0.4, col = c("#FF000080", "#0000FF80"))
circos.genomicDensity(bed_list[[1]], col = c("#FF000080"), track.height = 0.1)
circos.genomicDensity(bed_list[[2]], col = c("#0000FF80"), track.height = 0.1)
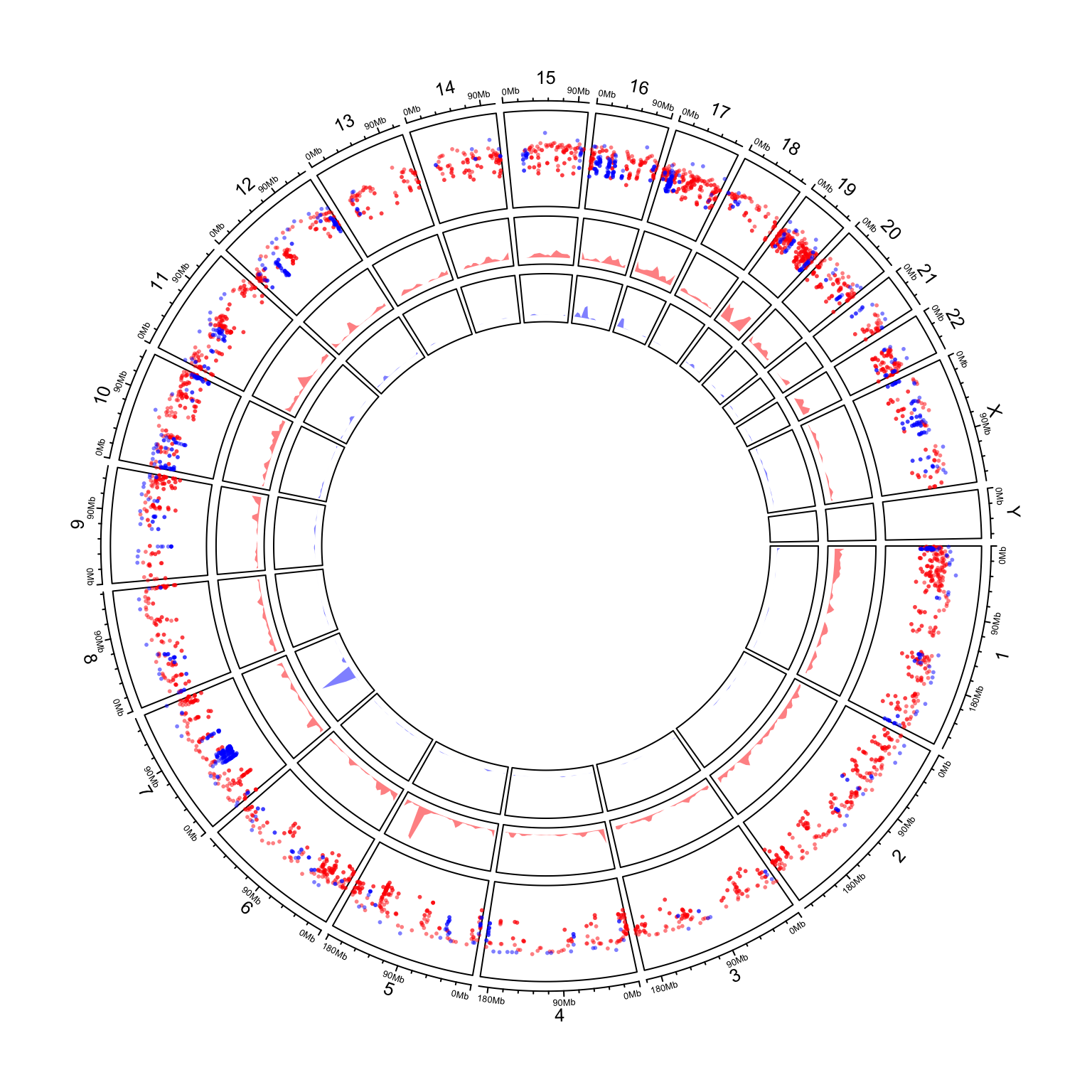
circos.clear()
Chord diagram.
mat = matrix(sample(1:100, 18, replace = TRUE), 3, 6)
rownames(mat) = letters[1:3]
colnames(mat) = LETTERS[1:6]
rn = rownames(mat)
cn = colnames(mat)
chordDiagram(mat)
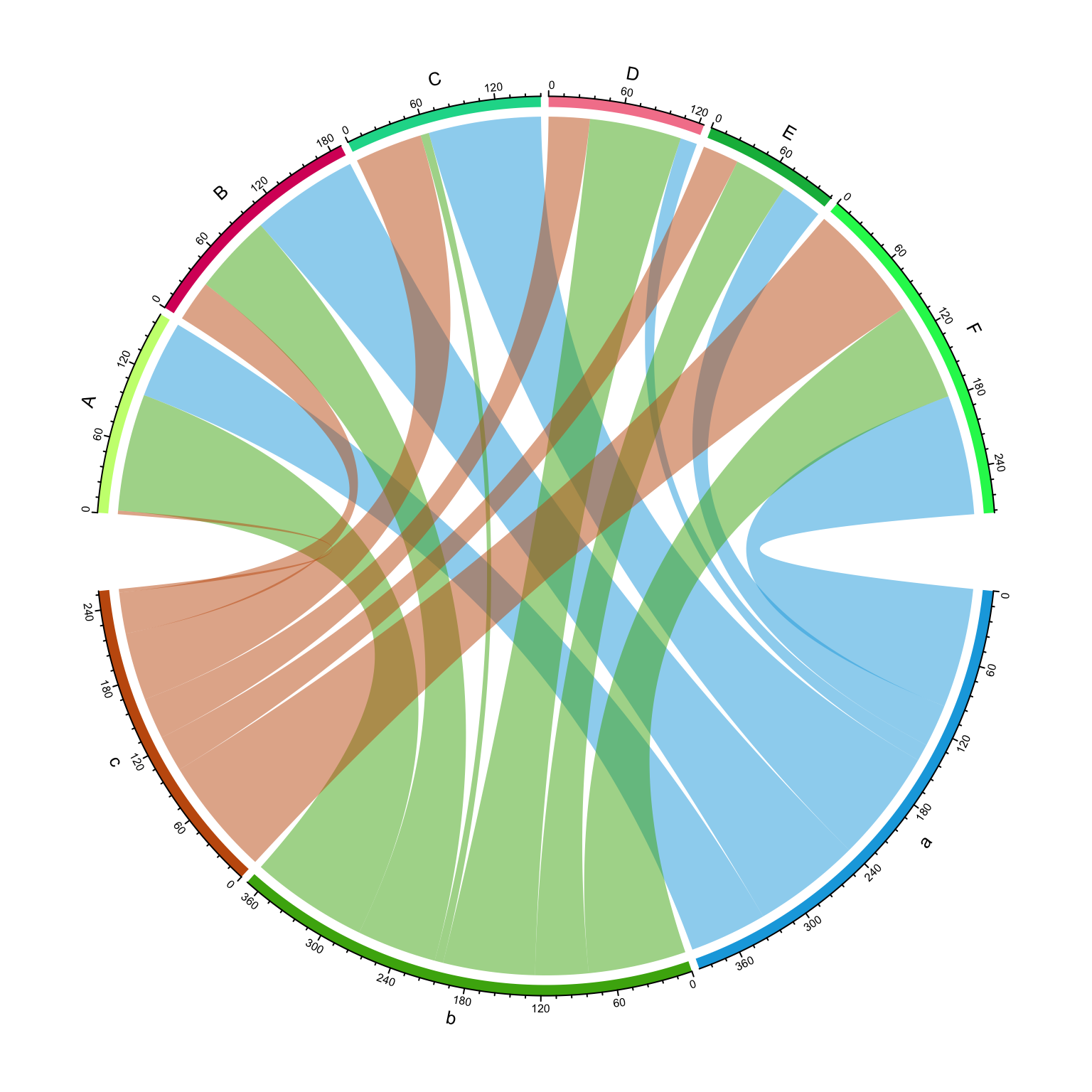
circos.clear()
More examples.
