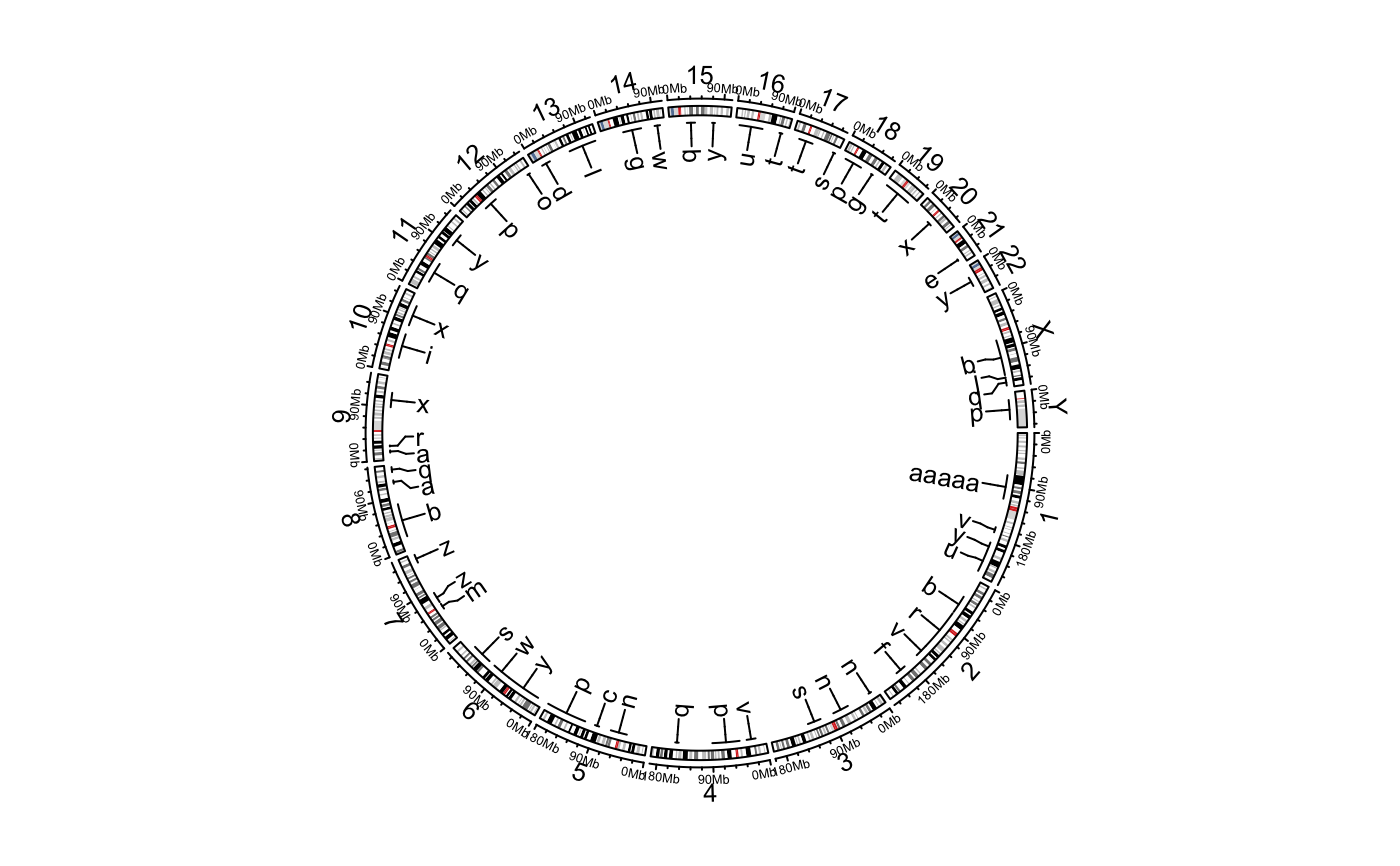
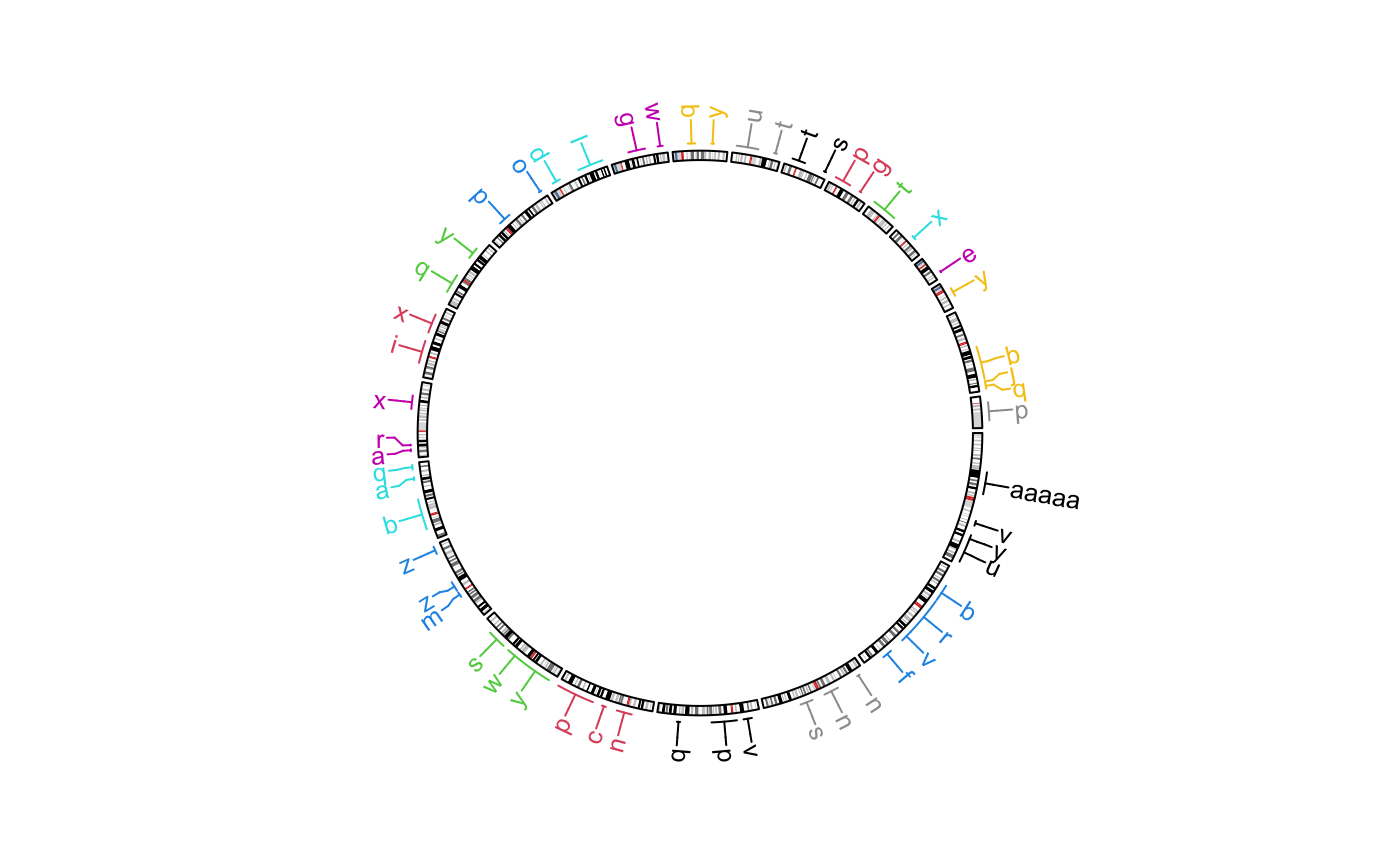
Add labels to specified genomic regions
circos.genomicLabels.RdAdd labels to specified genomic regions
circos.genomicLabels( bed, labels = NULL, labels.column = NULL, facing = "clockwise", niceFacing = TRUE, col = par("col"), cex = 0.8, font = par("font"), padding = 0.4, connection_height = mm_h(5), line_col = par("col"), line_lwd = par("lwd"), line_lty = par("lty"), labels_height = min(c(cm_h(1.5), max(strwidth(labels, cex = cex, font = font)))), side = c("inside", "outside"), track.margin = circos.par("track.margin"))
Arguments
| bed | A data frame in bed format. |
|---|---|
| labels | A vector of labels corresponding to rows in |
| labels.column | If the label column is already in |
| facing | fFacing of the labels. The value can only be |
| niceFacing | Whether automatically adjust the facing of the labels. |
| col | Color for the labels. |
| cex | Aize of the labels. |
| font | Font of the labels. |
| padding | Padding of the labels, the value is the ratio to the height of the label. |
| connection_height | Height of the connection track. |
| line_col | Color for the connection lines. |
| line_lwd | Line width for the connection lines. |
| line_lty | Line type for the connectioin lines. |
| labels_height | Height of the labels track. |
| side | Side of the labels track, is it in the inside of the track where the regions are marked? |
| track.margin | Bottom and top margins. |
Details
The function adds labels for the specified regions. The positions of labels are arranged so that they are not overlapping to each other.
This function creates two tracks, one for the connection lines and one for the labels.
See also
Examples
# \donttest{ circos.initializeWithIdeogram()bed = generateRandomBed(nr = 50, fun = function(k) sample(letters, k, replace = TRUE)) bed[1, 4] = "aaaaa" circos.genomicLabels(bed, labels.column = 4, side = "inside")circos.genomicLabels(bed, labels.column = 4, side = "outside", col = as.numeric(factor(bed[[1]])), line_col = as.numeric(factor(bed[[1]])))circos.clear() # }