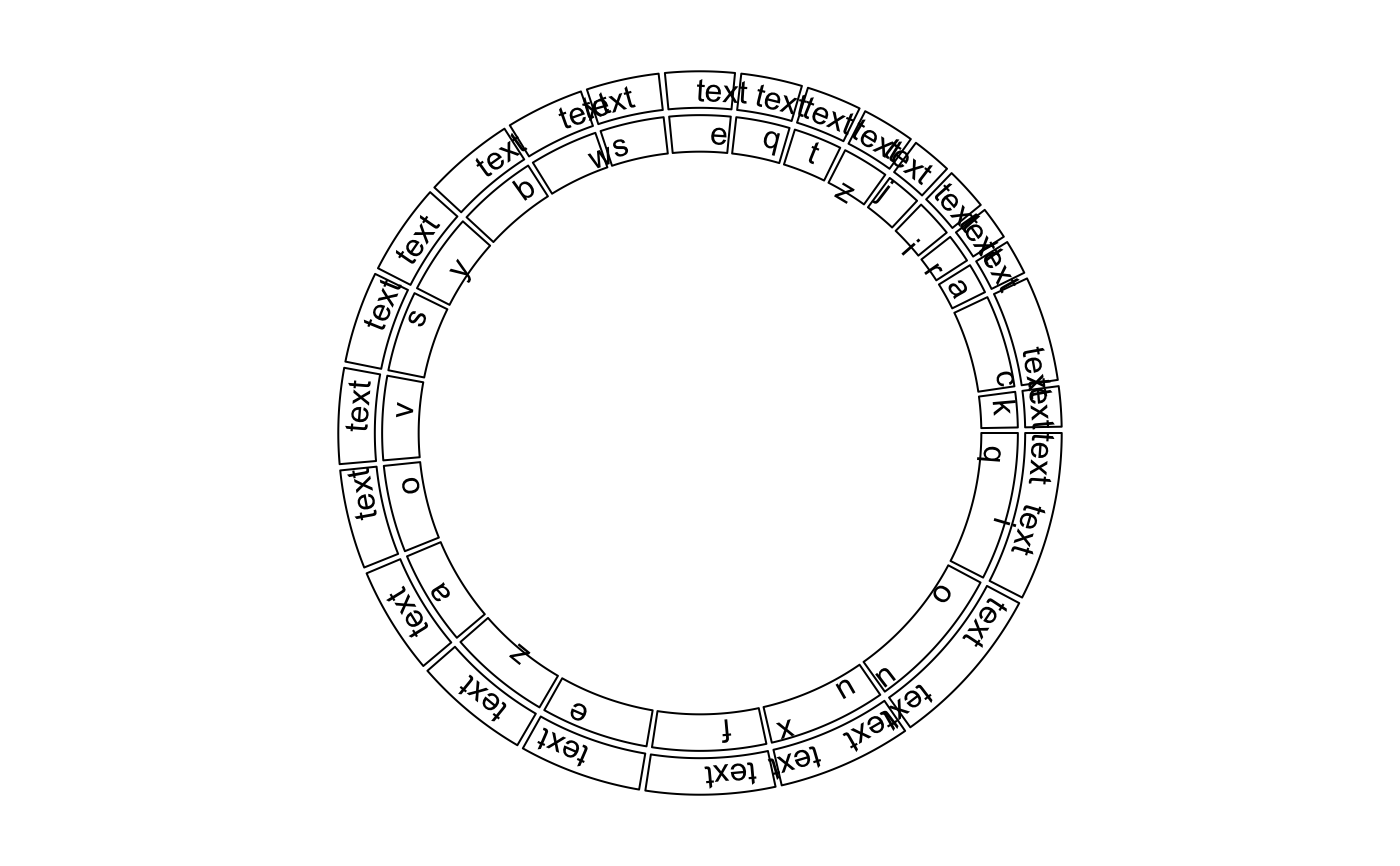
Draw text in a cell, specifically for genomic graphics
circos.genomicText.RdDraw text in a cell, specifically for genomic graphics
circos.genomicText( region, value = NULL, y = NULL, labels = NULL, labels.column = NULL, numeric.column = NULL, sector.index = get.current.sector.index(), track.index = get.current.track.index(), posTransform = NULL, direction = NULL, facing = "inside", niceFacing = FALSE, adj = par("adj"), cex = 1, col = "black", font = par("font"), padding = 0, extend = 0, align_to = "region", ...)
Arguments
| region | A data frame contains 2 column which correspond to start positions and end positions. |
|---|---|
| value | A data frame contains values and other information. |
| y | A vector or a single value indicating position of text. |
| labels | Labels of text corresponding to each genomic positions. |
| labels.column | If labels are in |
| numeric.column | Which column in |
| sector.index | Index of sector. |
| track.index | Index of track. |
| posTransform | Self-defined function to transform genomic positions, see |
| facing | Passing to |
| niceFacing | Should the facing of text be adjusted to fit human eyes? |
| direction | Deprecated, use |
| adj | Pass to |
| cex | Pass to |
| col | Pass to |
| font | Pass to |
| padding | pass to |
| extend | pass to |
| align_to | pass to |
| ... | Mysterious parameters. |
Details
The function is a low-level graphical function and usually is put in panel.fun when using circos.genomicTrack.
Examples
circos.par("track.height" = 0.1, cell.padding = c(0, 0, 0, 0)) circos.initializeWithIdeogram(plotType = NULL)bed = generateRandomBed(nr = 20) circos.genomicTrack(bed, ylim = c(0, 1), panel.fun = function(region, value, ...) { circos.genomicText(region, value, y = 0.5, labels = "text", ...) })bed = cbind(bed, sample(letters, nrow(bed), replace = TRUE)) circos.genomicTrack(bed, panel.fun = function(region, value, ...) { circos.genomicText(region, value, labels.column = 2, ...) })