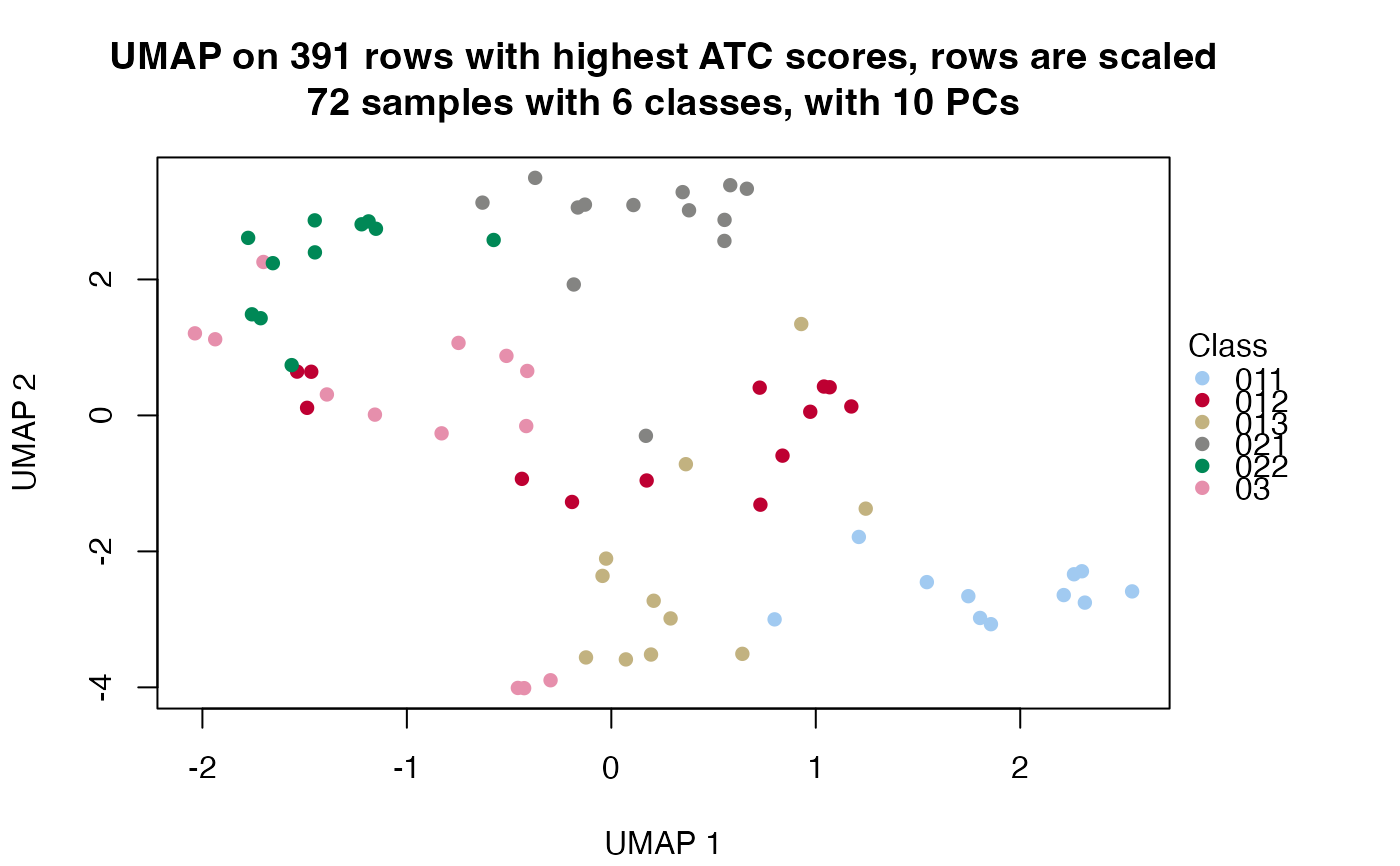
Visualize columns after dimension reduction
dimension_reduction-HierarchicalPartition-method.RdVisualize columns after dimension reduction
# S4 method for HierarchicalPartition
dimension_reduction(object, merge_node = merge_node_param(),
parent_node, top_n = NULL, top_value_method = object@list[[1]]@top_value_method,
method = c("PCA", "MDS", "t-SNE", "UMAP"), color_by = NULL,
scale_rows = object@list[[1]]@scale_rows, verbose = TRUE, ...)Arguments
- object
A
HierarchicalPartition-classobject.- merge_node
Parameters to merge sub-dendrograms, see
merge_node_param.- top_n
Top n rows to use. By default it uses all rows in the original matrix.
- top_value_method
Which top-value method to use.
- parent_node
Parent node. If it is set, the function call is identical to
dimension_reduction(object[parent_node])- method
Which method to reduce the dimension of the data.
MDSusescmdscale,PCAusesprcomp.t-SNEusesRtsne.UMAPusesumap.- color_by
If annotation table is set, an annotation name can be set here.
- scale_rows
Whether to perform scaling on matrix rows.
- verbose
Whether print messages.
- ...
Other arguments passed to
dimension_reduction,ConsensusPartition-method.
Details
The class IDs are extract at depth.
Value
No value is returned.
Examples
data(golub_cola_rh)
dimension_reduction(golub_cola_rh)
#> use UMAP