When making heatmaps, we always have many additionally grouping information
for the samples, sush as subtypes, phenotypes. To emphasize the difference
between groups, we might want to only perform clustering on group level, while
not on the complete dataset. For these scenarios, ComplexHeatmap has two
functions: cluster_between_groups() and cluster_within_group(). The two
functions all perform clustering on group level. The difference is, in
cluster_between_groups(), inside each group, the order of samples is
unchanged, while in cluster_within_group(), samples in each group are still
clustered.
I demonstrate the use of the two functions with a simple random matrix. In the
first plot, cluster_between_groups() was applied. The column names
correspond to the original order in the matrix, so you can observe whether the
columns are reordered or not.
library(ComplexHeatmap)
set.seed(372)
m = matrix(rnorm(120), nc = 12)
colnames(m) = 1:12
fa = rep(c("a", "b", "c"), times = c(2, 4, 6))
fa_col = c("a" = 2, "b" = 3, "c" = 4)
dend1 = cluster_between_groups(m, fa)
Heatmap(m, cluster_columns = dend1, column_split = 3,
row_title = "cluster_between_groups",
top_annotation = HeatmapAnnotation(foo = fa, col = list(foo = fa_col)))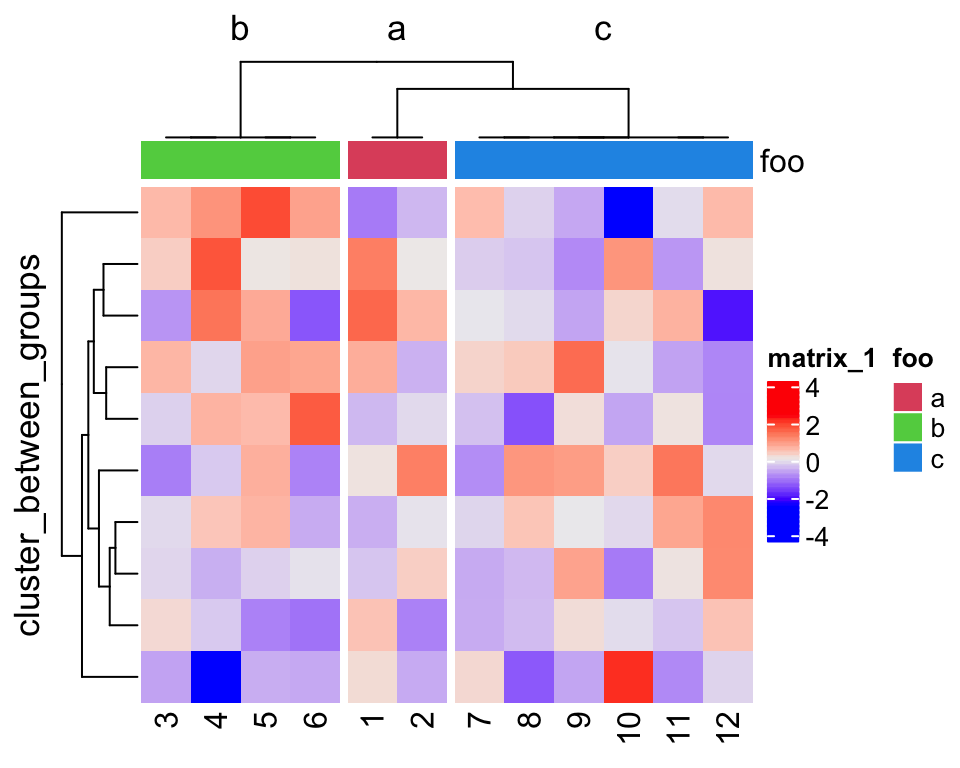
In the second plot, cluster_within_group() was applied. As you can see, columns
are also reordered inside each group.
dend2 = cluster_within_group(m, fa)
Heatmap(m, cluster_columns = dend2, column_split = 3,
row_title = "cluster_within_group",
top_annotation = HeatmapAnnotation(foo = fa, col = list(foo = fa_col)))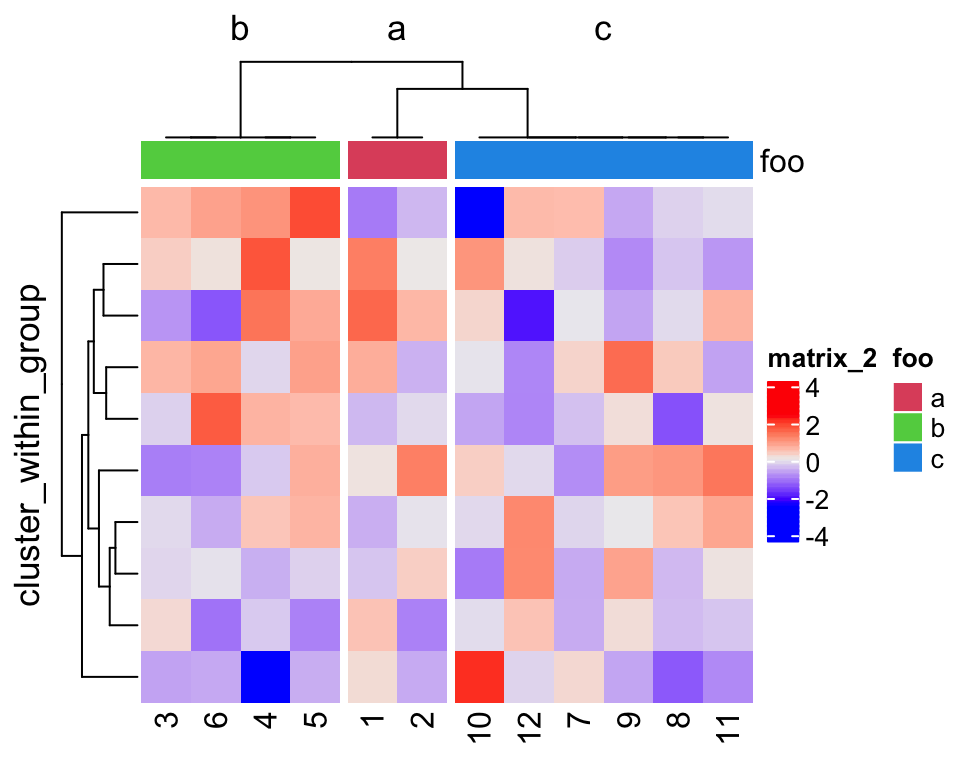
Note, they also work on heatmap rows. Please update to the current GitHub version to use these two functions.
Finally, thanks @crazyhottommy for providing this interesting use case.