library(simplifyEnrichment)To perform GO clustering with the simplifyEnrichment package, the first step
is to calculate GO similarities with the function GO_similarity(). The semantic similarity
depends on the genes annotated to GO terms, thus, an OrgDb package is needed for
the use of GO_similarity(). For example, for mouse:
GO_similarity(go_id, db = "org.Mm.eg.db")On Bioconductor, there are the following OrgDb packages that can be used with GO_similarity().
| Package | Organism |
|---|---|
| org.Hs.eg.db | Human |
| org.Mm.eg.db | Mouse |
| org.Rn.eg.db | Rat |
| org.Dm.eg.db | Fly |
| org.At.tair.db | Arabidopsis |
| org.Sc.sgd.db | Yeast |
| org.Dr.eg.db | Zebrafish |
| org.Ce.eg.db | Worm |
| org.Bt.eg.db | Bovine |
| org.Gg.eg.db | Chicken |
| org.Ss.eg.db | Pig |
| org.Mmu.eg.db | Rhesus |
| org.Cf.eg.db | Canine |
| org.EcK12.eg.db | E coli strain K12 |
| org.Xl.eg.db | Xenopus |
| org.Ag.eg.db | Anopheles |
| org.Pt.eg.db | Chimp |
| org.EcSakai.eg.db | E coli strain Sakai |
| org.Mxanthus.db | Myxococcus xanthus DK 1622 |
But what if there is no such Bioconductor package for your organism? Don’t worry, there are more resources provided on the Bioconductor cloud, where the data can be easily retrieved with the AnnotationHub package.
To use AnnotationHub, first we need to create an AnnotationHub object. In this step, a local cache
folder will be created and the same data will be directly read from the cache.
library(AnnotationHub)
ah = AnnotationHub()
ah## AnnotationHub with 69797 records
## # snapshotDate(): 2022-10-31
## # $dataprovider: Ensembl, BroadInstitute, UCSC, ftp://ftp.ncbi.nlm.nih.gov/g...
## # $species: Homo sapiens, Mus musculus, Drosophila melanogaster, Bos taurus,...
## # $rdataclass: GRanges, TwoBitFile, BigWigFile, EnsDb, Rle, OrgDb, ChainFile...
## # additional mcols(): taxonomyid, genome, description,
## # coordinate_1_based, maintainer, rdatadateadded, preparerclass, tags,
## # rdatapath, sourceurl, sourcetype
## # retrieve records with, e.g., 'object[["AH5012"]]'
##
##
## AH5012 |
## AH5013 |
## AH5014 |
## AH5015 |
## AH5016 |
## ...
## AH111329 |
## AH111330 |
## AH111331 |
## AH111332 |
## AH111334 |
## title
## AH5012 Chromosome Band
## AH5013 STS Markers
## AH5014 FISH Clones
## AH5015 Recomb Rate
## AH5016 ENCODE Pilot
## ... ...
## AH111329 Zonotrichia_albicollis.Zonotrichia_albicollis-1.0.1.109.abinitio...
## AH111330 Zonotrichia_albicollis.Zonotrichia_albicollis-1.0.1.109.gtf
## AH111331 Zosterops_lateralis_melanops.ASM128173v1.109.abinitio.gtf
## AH111332 Zosterops_lateralis_melanops.ASM128173v1.109.gtf
## AH111334 MassBank CompDb for release 2022.12.1ah contains summary of all available datasets on the cloud. To download an OrgDb object for a specific
organism, we can pre-filter ah to let it only restrict on the OrgDb data type.
ah = ah[ah$rdataclass == "OrgDb"]
ah## AnnotationHub with 1871 records
## # snapshotDate(): 2022-10-31
## # $dataprovider: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/
## # $species: Escherichia coli, greater Indian_fruit_bat, Zootoca vivipara, Zo...
## # $rdataclass: OrgDb
## # additional mcols(): taxonomyid, genome, description,
## # coordinate_1_based, maintainer, rdatadateadded, preparerclass, tags,
## # rdatapath, sourceurl, sourcetype
## # retrieve records with, e.g., 'object[["AH107050"]]'
##
## title
## AH107050 | org.Ag.eg.db.sqlite
## AH107051 | org.At.tair.db.sqlite
## AH107052 | org.Bt.eg.db.sqlite
## AH107053 | org.Cf.eg.db.sqlite
## AH107054 | org.Gg.eg.db.sqlite
## ... ...
## AH109234 | org.Rhizoctonia_praticola.eg.sqlite
## AH109235 | org.Rhizoctonia_solani.eg.sqlite
## AH109236 | org.Heterostelium_album_PN500.eg.sqlite
## AH109237 | org.Heterostelium_pallidum_PN500.eg.sqlite
## AH109238 | org.Polysphondylium_pallidum_PN500.eg.sqliteStill, you can see there are more than 1800 organisms with their OrgDb data available on the Bioconductor cloud.
So it is very likely you can find your organism there.
Use mcols() to get a more informative description of all OrgDb records. In the following table,
IDs as row names (e.g. "AH5012") will be used to retrieve the corresponding dataset from the cloud.
head(mcols(ah))## DataFrame with 6 rows and 15 columns
## title dataprovider species
## <character> <character> <character>
## AH107050 org.Ag.eg.db.sqlite ftp://ftp.ncbi.nlm.n.. Anopheles gambiae
## AH107051 org.At.tair.db.sqlite ftp://ftp.ncbi.nlm.n.. Arabidopsis thaliana
## AH107052 org.Bt.eg.db.sqlite ftp://ftp.ncbi.nlm.n.. Bos taurus
## AH107053 org.Cf.eg.db.sqlite ftp://ftp.ncbi.nlm.n.. Canis familiaris
## AH107054 org.Gg.eg.db.sqlite ftp://ftp.ncbi.nlm.n.. Gallus gallus
## AH107055 org.Pt.eg.db.sqlite ftp://ftp.ncbi.nlm.n.. Pan troglodytes
## taxonomyid genome description coordinate_1_based
## <integer> <character> <character> <integer>
## AH107050 180454 NCBI genomes NCBI gene ID based a.. 1
## AH107051 3702 NCBI genomes NCBI gene ID based a.. 1
## AH107052 9913 NCBI genomes NCBI gene ID based a.. 1
## AH107053 9615 NCBI genomes NCBI gene ID based a.. 1
## AH107054 9031 NCBI genomes NCBI gene ID based a.. 1
## AH107055 9598 NCBI genomes NCBI gene ID based a.. 1
## maintainer rdatadateadded preparerclass
## <character> <character> <character>
## AH107050 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## AH107051 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## AH107052 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## AH107053 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## AH107054 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## AH107055 Bioconductor Maintai.. 2022-09-29 OrgDbFromPkgsImportP..
## tags rdataclass rdatapath
## <AsIs> <character> <character>
## AH107050 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## AH107051 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## AH107052 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## AH107053 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## AH107054 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## AH107055 NCBI,Gene,Annotation OrgDb ncbi/standard/3.16/o..
## sourceurl sourcetype
## <character> <character>
## AH107050 ftp://ftp.ncbi.nlm.n.. NCBI/ensembl
## AH107051 ftp://ftp.ncbi.nlm.n.. NCBI/ensembl
## AH107052 ftp://ftp.ncbi.nlm.n.. NCBI/ensembl
## AH107053 ftp://ftp.ncbi.nlm.n.. NCBI/ensembl
## AH107054 ftp://ftp.ncbi.nlm.n.. NCBI/ensembl
## AH107055 ftp://ftp.ncbi.nlm.n.. NCBI/ensemblThere are two ways to search for the specific organism. First you can directly search in the
meta columns of ah (suggested columns are species, taxonomyid, description). For example:
meta = mcols(ah)
rownames(meta)[ which(meta$species == "Bos taurus") ]## [1] "AH107052"rownames(meta)[ which(meta$taxonomyid == 9031) ]## [1] "AH107054"Second, you can use the query() function by providing a list of keywords. To search for a specific
organism, I suggest to use its Latin name. In the next example, I search for the data for dolphins (Latin name is “Tursiops truncatus”).
query(ah, "Tursiops truncatus")## AnnotationHub with 1 record
## # snapshotDate(): 2022-10-31
## # names(): AH108106
## # $dataprovider: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/
## # $species: Tursiops truncatus
## # $rdataclass: OrgDb
## # $rdatadateadded: 2022-10-26
## # $title: org.Tursiops_truncatus.eg.sqlite
## # $description: NCBI gene ID based annotations about Tursiops truncatus
## # $taxonomyid: 9739
## # $genome: NCBI genomes
## # $sourcetype: NCBI/UniProt
## # $sourceurl: ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/, ftp://ftp.uniprot.org/p...
## # $sourcesize: NA
## # $tags: c("NCBI", "Gene", "Annotation")
## # retrieve record with 'object[["AH108106"]]'The Hub ID is "AH108106". Then we can retrieve the OrgDb object with ah[["AH108106"]]:
orgdb = ah[["AH108106"]]
orgdb## OrgDb object:
## | DBSCHEMAVERSION: 2.1
## | DBSCHEMA: NOSCHEMA_DB
## | ORGANISM: Tursiops truncatus
## | SPECIES: Tursiops truncatus
## | CENTRALID: GID
## | Taxonomy ID: 9739
## | Db type: OrgDb
## | Supporting package: AnnotationDbiWe can check the supported columns in the object orgdb:
columns(orgdb)## [1] "ACCNUM" "ALIAS" "CHR" "ENSEMBL" "ENTREZID"
## [6] "EVIDENCE" "EVIDENCEALL" "GENENAME" "GID" "GO"
## [11] "GOALL" "ONTOLOGY" "ONTOLOGYALL" "PMID" "REFSEQ"
## [16] "SYMBOL"In the next section of code, I will demonstrate how to perform simplifyEnrichment analysis on dolphnins. First I generate a random list of GO IDs.
all_GO_ids = select(orgdb, keys(orgdb, keytype = "GO"), c("GO", "ONTOLOGY"), keytype = "GO")
all_GO_ids = all_GO_ids[all_GO_ids$ONTOLOGY == "BP", ]
set.seed(666)
go_id = sample(all_GO_ids$GO, 500)
head(go_id)## [1] "GO:0034969" "GO:0007223" "GO:0070816" "GO:0010845" "GO:1905259"
## [6] "GO:0071300"Next I calculate the GO similarities. This time, I directly set the db argument with the orgdb object.
mat = GO_similarity(go_id, db = orgdb)If you are not using the newest version of simplifyEnrichment, you need to also set ont = "BP" to get rid of errors:
mat = GO_similarity(go_id, db = orgdb, ont = "BP")And make the heatmap with word clouds.
simplifyGO(mat)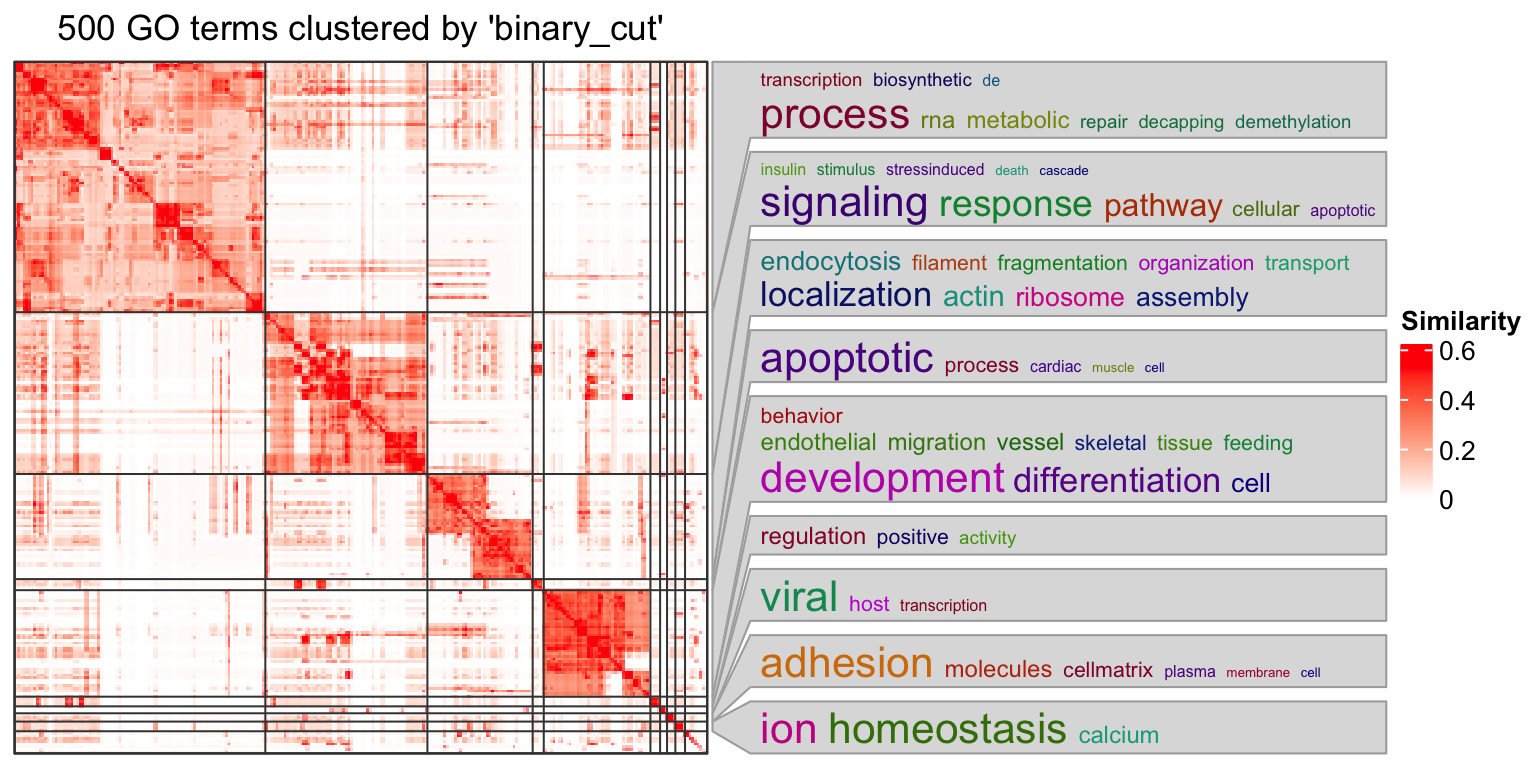
Session info
sessionInfo()## R version 4.2.0 (2022-04-22)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur/Monterey 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] C/UTF-8/C/C/C/C
##
## attached base packages:
## [1] stats4 grid stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] AnnotationDbi_1.60.0 IRanges_2.32.0 S4Vectors_0.36.1
## [4] Biobase_2.58.0 AnnotationHub_3.6.0 BiocFileCache_2.6.0
## [7] dbplyr_2.3.0 simplifyEnrichment_1.9.2 BiocGenerics_0.44.0
## [10] knitr_1.42
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-7 matrixStats_0.63.0
## [3] bit64_4.0.5 filelock_1.0.2
## [5] doParallel_1.0.17 RColorBrewer_1.1-3
## [7] httr_1.4.4 GenomeInfoDb_1.34.9
## [9] tools_4.2.0 bslib_0.4.2
## [11] utf8_1.2.3 R6_2.5.1
## [13] DBI_1.1.3 colorspace_2.1-0
## [15] GetoptLong_1.1.0 withr_2.5.0
## [17] tidyselect_1.2.0 proxyC_0.3.3
## [19] bit_4.0.5 curl_5.0.0
## [21] compiler_4.2.0 cli_3.6.0
## [23] Cairo_1.6-0 xml2_1.3.3
## [25] NLP_0.2-1 bookdown_0.32
## [27] slam_0.1-50 sass_0.4.5
## [29] tm_0.7-11 rappdirs_0.3.3
## [31] digest_0.6.31 rmarkdown_2.20
## [33] XVector_0.38.0 pkgconfig_2.0.3
## [35] htmltools_0.5.4 highr_0.10
## [37] fastmap_1.1.0 rlang_1.0.6
## [39] GlobalOptions_0.1.2 RSQLite_2.2.20
## [41] shiny_1.7.4 shape_1.4.6
## [43] jquerylib_0.1.4 generics_0.1.3
## [45] jsonlite_1.8.4 GOSemSim_2.24.0
## [47] dplyr_1.1.0 RCurl_1.98-1.10
## [49] magrittr_2.0.3 GO.db_3.16.0
## [51] GenomeInfoDbData_1.2.9 Matrix_1.5-3
## [53] Rcpp_1.0.10 fansi_1.0.4
## [55] lifecycle_1.0.3 yaml_2.3.7
## [57] zlibbioc_1.44.0 org.Hs.eg.db_3.16.0
## [59] blob_1.2.3 promises_1.2.0.1
## [61] parallel_4.2.0 crayon_1.5.2
## [63] lattice_0.20-45 Biostrings_2.66.0
## [65] circlize_0.4.16 KEGGREST_1.38.0
## [67] magick_2.7.3 ComplexHeatmap_2.15.2
## [69] pillar_1.8.1 rjson_0.2.21
## [71] codetools_0.2-19 glue_1.6.2
## [73] BiocVersion_3.16.0 evaluate_0.20
## [75] blogdown_1.16 BiocManager_1.30.20
## [77] RcppParallel_5.1.6 httpuv_1.6.8
## [79] png_0.1-8 vctrs_0.5.2
## [81] foreach_1.5.2 purrr_1.0.1
## [83] clue_0.3-64 assertthat_0.2.1
## [85] cachem_1.0.6 xfun_0.37
## [87] mime_0.12 xtable_1.8-4
## [89] later_1.3.0 tibble_3.1.8
## [91] iterators_1.0.14 memoise_2.0.1
## [93] cluster_2.1.4 ellipsis_0.3.2
## [95] interactiveDisplayBase_1.36.0