Get signatures rows
get_signatures-HierarchicalPartition-method.RdGet signatures rows
# S4 method for HierarchicalPartition
get_signatures(object, merge_node = merge_node_param(),
group_diff = object@param$group_diff,
row_km = NULL, diff_method = "Ftest", fdr_cutoff = object@param$fdr_cutoff,
scale_rows = object[1]@scale_rows,
anno = get_anno(object),
anno_col = get_anno_col(object),
show_column_names = FALSE, column_names_gp = gpar(fontsize = 8),
verbose = TRUE, plot = TRUE, seed = 888,
...)Arguments
- object
a
HierarchicalPartition-classobject.- merge_node
Parameters to merge sub-dendrograms, see
merge_node_param.- group_diff
Cutoff for the maximal difference between group means.
- row_km
Number of groups for performing k-means clustering on rows. By default it is automatically selected.
- diff_method
Methods to get rows which are significantly different between subgroups.
- fdr_cutoff
Cutoff for FDR of the difference test between subgroups.
- scale_rows
whether apply row scaling when making the heatmap.
- anno
a data frame of annotations for the original matrix columns. By default it uses the annotations specified in
hierarchical_partition.- anno_col
a list of colors (color is defined as a named vector) for the annotations. If
annois a data frame,anno_colshould be a named list where names correspond to the column names inanno.- show_column_names
whether show column names in the heatmap.
- column_names_gp
Graphic parameters for column names.
- verbose
whether to print messages.
- plot
whether to make the plot.
- seed
Random seed.
- ...
other arguments pass to
get_signatures,ConsensusPartition-method.
Details
The function calls get_signatures,ConsensusPartition-method to find signatures at
each node of the partition hierarchy.
Value
A data frame with more than two columns:
which_row:row index corresponding to the original matrix.
km:the k-means groups if
row_kmis set.- other_columns:
the mean value (depending rows are scaled or not) in each subgroup.
Examples
# \donttest{
data(golub_cola_rh)
tb = get_signatures(golub_cola_rh)
#> * get signatures at node 0 with 3 subgroups.
#> * 71/72 samples (in 3 classes) remain after filtering by silhouette (>= 0.5).
#> * cache hash: 69feee4d94f63bf432b51d8e53ece52c (seed 888).
#> * calculating row difference between subgroups by Ftest.
#> * get signatures at node 01 with 3 subgroups.
#> * 34/35 samples (in 3 classes) remain after filtering by silhouette (>= 0.5).
#> * cache hash: 2998831f64132aeaf6eb44c6e6912575 (seed 888).
#> * calculating row difference between subgroups by Ftest.
#> * get signatures at node 02 with 2 subgroups.
#> * 24/24 samples (in 2 classes) remain after filtering by silhouette (>= 0.5).
#> * cache hash: ea242e8e77a1cc217cae91637d6d95f6 (seed 888).
#> * calculating row difference between subgroups by Ftest.
#> * split rows into 4 groups by k-means clustering.
#> * found 2350 signatures (57.1%).
#> * randomly sample 2000 rows from 2350 total rows.
#> * making heatmaps for signatures
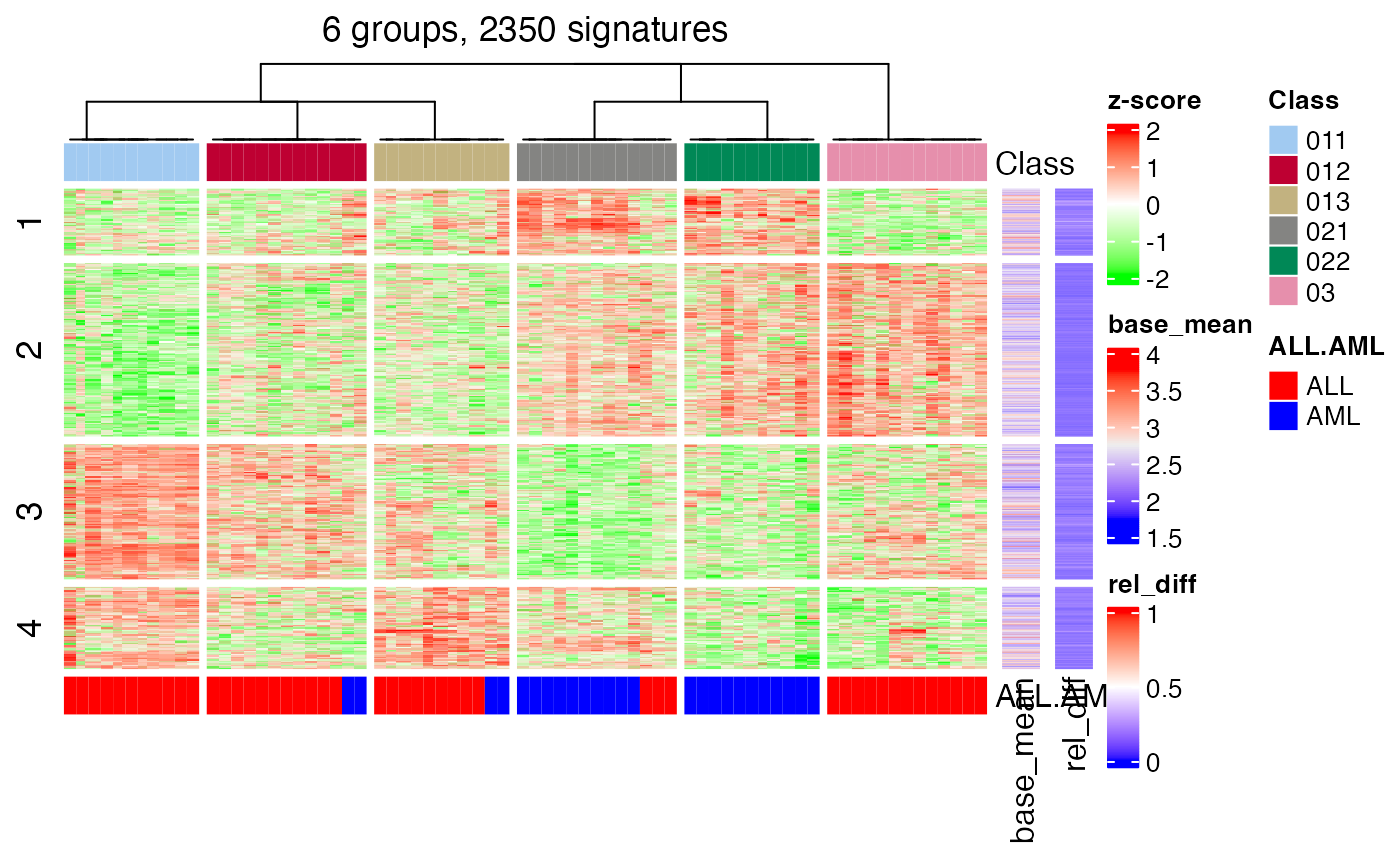 head(tb)
#> which_row is_sig_0 is_sig_01 is_sig_02 mean_011 mean_012 mean_013 mean_021
#> 1 2 TRUE FALSE FALSE 1.641389 1.845958 1.762456 2.091835
#> 2 3 TRUE FALSE FALSE 2.184402 2.206852 2.183741 2.171343
#> 3 11 TRUE FALSE FALSE 2.288716 2.333027 2.427159 2.600189
#> 4 12 TRUE FALSE FALSE 2.269837 2.357288 2.565022 2.690489
#> 5 13 TRUE FALSE FALSE 2.424987 3.011391 2.795872 3.151072
#> 6 14 TRUE FALSE FALSE 3.035862 3.445460 3.269206 3.489674
#> mean_022 mean_03 group_diff scaled_mean_011 scaled_mean_012 scaled_mean_013
#> 1 2.068923 2.003520 0.4504458 -0.8295840 -0.1950825 -0.4540758
#> 2 2.346207 2.386800 0.2154572 -0.2994006 -0.1924667 -0.3025460
#> 3 2.445832 2.366862 0.3114738 -0.6207227 -0.3980173 0.0750845
#> 4 2.382775 2.265742 0.4247472 -0.6046877 -0.2598623 0.5592590
#> 5 2.473630 3.416868 0.9918814 -0.6637468 0.1468631 -0.1510570
#> 6 3.232134 3.622108 0.5862460 -0.8241242 0.2069714 -0.2367195
#> scaled_mean_021 scaled_mean_022 scaled_mean_03 group_diff_scaled km
#> 1 0.5675420 0.4964768 0.2936185 1.397126 2
#> 2 -0.3616000 0.4712822 0.6646289 1.026229 2
#> 3 0.9447291 0.1689359 -0.2279637 1.565452 1
#> 4 1.0539864 -0.1593632 -0.6208386 1.674825 1
#> 5 0.3399502 -0.5965052 0.7073713 1.371118 2
#> 6 0.3182753 -0.3300409 0.6516556 1.475780 2
# }
head(tb)
#> which_row is_sig_0 is_sig_01 is_sig_02 mean_011 mean_012 mean_013 mean_021
#> 1 2 TRUE FALSE FALSE 1.641389 1.845958 1.762456 2.091835
#> 2 3 TRUE FALSE FALSE 2.184402 2.206852 2.183741 2.171343
#> 3 11 TRUE FALSE FALSE 2.288716 2.333027 2.427159 2.600189
#> 4 12 TRUE FALSE FALSE 2.269837 2.357288 2.565022 2.690489
#> 5 13 TRUE FALSE FALSE 2.424987 3.011391 2.795872 3.151072
#> 6 14 TRUE FALSE FALSE 3.035862 3.445460 3.269206 3.489674
#> mean_022 mean_03 group_diff scaled_mean_011 scaled_mean_012 scaled_mean_013
#> 1 2.068923 2.003520 0.4504458 -0.8295840 -0.1950825 -0.4540758
#> 2 2.346207 2.386800 0.2154572 -0.2994006 -0.1924667 -0.3025460
#> 3 2.445832 2.366862 0.3114738 -0.6207227 -0.3980173 0.0750845
#> 4 2.382775 2.265742 0.4247472 -0.6046877 -0.2598623 0.5592590
#> 5 2.473630 3.416868 0.9918814 -0.6637468 0.1468631 -0.1510570
#> 6 3.232134 3.622108 0.5862460 -0.8241242 0.2069714 -0.2367195
#> scaled_mean_021 scaled_mean_022 scaled_mean_03 group_diff_scaled km
#> 1 0.5675420 0.4964768 0.2936185 1.397126 2
#> 2 -0.3616000 0.4712822 0.6646289 1.026229 2
#> 3 0.9447291 0.1689359 -0.2279637 1.565452 1
#> 4 1.0539864 -0.1593632 -0.6208386 1.674825 1
#> 5 0.3399502 -0.5965052 0.7073713 1.371118 2
#> 6 0.3182753 -0.3300409 0.6516556 1.475780 2
# }