Predict classes for new samples based on cola classification
predict_classes-ConsensusPartition-method.RdPredict classes for new samples based on cola classification
# S4 method for ConsensusPartition
predict_classes(object, k, mat,
silhouette_cutoff = 0.5,
fdr_cutoff = cola_opt$fdr_cutoff,
group_diff = cola_opt$group_diff,
scale_rows = object@scale_rows,
diff_method = "Ftest",
method = "centroid",
dist_method = c("euclidean", "correlation", "cosine"), nperm = 1000,
p_cutoff = 0.05, plot = TRUE, col_fun = NULL,
split_by_sigatures = FALSE, force = FALSE,
verbose = TRUE, help = TRUE, prefix = "",
mc.cores = 1, cores = mc.cores)Arguments
- object
A
ConsensusPartition-classobject.- k
Number of subgroups to get the classifications.
- mat
The new matrix where the sample classes are going to be predicted. The number of rows should be the same as the original matrix for cola analysis (also make sure the row orders are the same). Be careful that the scaling of
matshould be the same as that applied in cola analysis.- silhouette_cutoff
Send to
get_signatures,ConsensusPartition-methodfor determining signatures.- fdr_cutoff
Send to
get_signatures,ConsensusPartition-methodfor determining signatures.- group_diff
Send to
get_signatures,ConsensusPartition-methodfor determining signatures.- scale_rows
Send to
get_signatures,ConsensusPartition-methodfor determining signatures.- diff_method
Send to
get_signatures,ConsensusPartition-methodfor determining signatures.- method
Method for predicting class labels. Possible values are "centroid", "svm" and "randomForest".
- dist_method
Distance method. Value should be "euclidean", "correlation" or "cosine". Send to
predict_classes,matrix-method.- nperm
Number of permutatinos. It is used when
dist_methodis set to "euclidean" or "cosine". Send topredict_classes,matrix-method.- p_cutoff
Cutoff for the p-values for determining class assignment. Send to
predict_classes,matrix-method.- plot
Whether to draw the plot that visualizes the process of prediction. Send to
predict_classes,matrix-method.- col_fun
A color mapping function generated from
colorRamp2. It is set to both heatmaps.- split_by_sigatures
Should the heatmaps be split based on k-means on the main heatmap, or on the patterns of the signature heatmap.
- force
If the value is
TRUEand whenget_signatures,ConsensusPartition-methodinternally failed, top 1000 rows with the highest between-group mean difference are used for constructing the signature centroid matrix. It is basically used internally.- verbose
Whether to print messages. Send to
predict_classes,matrix-method.- help
Whether to print help messages.
- prefix
Used internally.
- mc.cores
Number of cores. This argument will be removed in future versions.
- cores
Number of cores, or a
clusterobject returned bymakeCluster.
Details
The prediction is based on the signature centroid matrix from cola classification. The processes are as follows:
1. For the provided ConsensusPartition-class object and a selected k, the signatures that discriminate classes
are extracted by get_signatures,ConsensusPartition-method. If number of signatures is more than 2000, only 2000 signatures are randomly sampled.
2. The signature centroid matrix is a k-column matrix where each column is the centroid of samples in the corresponding
class, i.e. the mean across samples. If rows were scaled in cola analysis, the signature centroid matrix is the mean of scaled
values and vise versa. Please note the samples with silhouette score less than silhouette_cutoff are removed
for calculating the centroids.
3. With the signature centroid matrix and the new matrix, it calls predict_classes,matrix-method to perform the prediction.
Please see more details of the prediction on that help page. Please note, the scales of the new matrix should be the same as the matrix
used for cola analysis.
Value
A data frame with two columns: the class labels (in numeric) and the corresponding p-values.
See also
predict_classes,matrix-method that predicts the classes for new samples.
Examples
# \donttest{
data(golub_cola)
res = golub_cola["ATC:skmeans"]
mat = get_matrix(res)
# note scaling should be applied here because the matrix was scaled in the cola analysis
mat2 = t(scale(t(mat)))
cl = predict_classes(res, k = 3, mat2)
#> The matrix has been scaled in cola analysis, thus the new matrix should
#> also be scaled with the same method ('z-score'). Please double check.
#> Set `help = FALSE` to suppress this message.
#>
#> * take top 500/2058 most significant signatures for prediction.
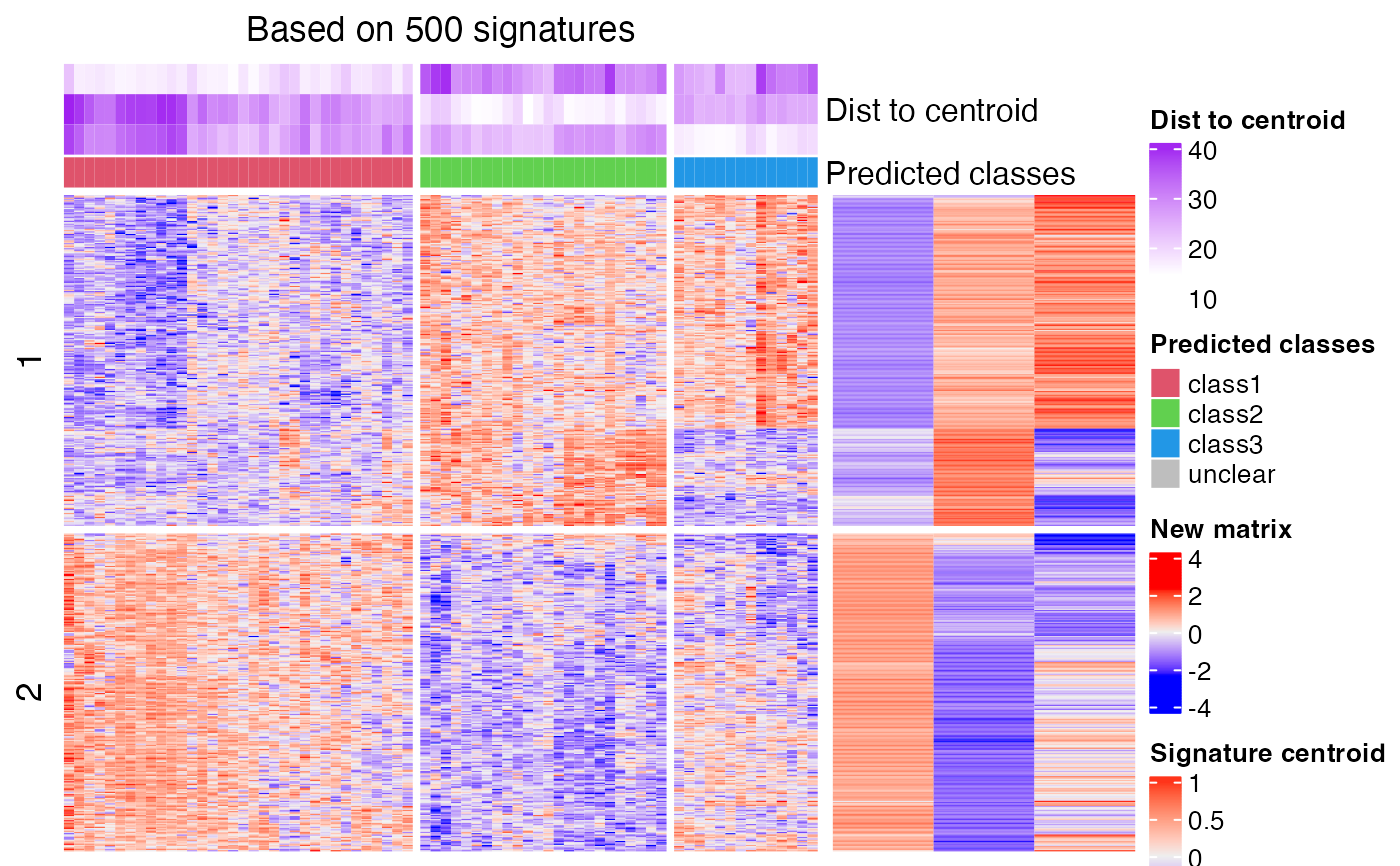 # compare the real classification and the predicted classification
data.frame(cola_class = get_classes(res, k = 3)[, "class"],
predicted = cl[, "class"])
#> cola_class predicted
#> 1 3 3
#> 2 3 3
#> 3 1 1
#> 4 1 1
#> 5 1 1
#> 6 3 3
#> 7 1 1
#> 8 1 1
#> 9 1 1
#> 10 1 1
#> 11 1 1
#> 12 1 1
#> 13 1 1
#> 14 1 1
#> 15 1 1
#> 16 1 1
#> 17 1 1
#> 18 3 3
#> 19 3 3
#> 20 1 1
#> 21 2 2
#> 22 2 2
#> 23 2 2
#> 24 2 2
#> 25 1 1
#> 26 2 2
#> 27 2 2
#> 28 1 1
#> 29 2 2
#> 30 2 2
#> 31 1 1
#> 32 2 2
#> 33 2 2
#> 34 2 2
#> 35 3 3
#> 36 1 1
#> 37 3 3
#> 38 3 3
#> 39 1 1
#> 40 3 3
#> 41 3 3
#> 42 3 3
#> 43 1 1
#> 44 1 1
#> 45 1 1
#> 46 2 2
#> 47 1 1
#> 48 1 1
#> 49 1 1
#> 50 1 1
#> 51 1 1
#> 52 3 3
#> 53 1 1
#> 54 1 1
#> 55 1 1
#> 56 2 2
#> 57 3 3
#> 58 1 1
#> 59 2 2
#> 60 1 1
#> 61 3 3
#> 62 2 2
#> 63 2 2
#> 64 2 2
#> 65 2 2
#> 66 2 2
#> 67 2 2
#> 68 1 1
#> 69 2 2
#> 70 2 2
#> 71 2 2
#> 72 2 2
# change to correlation method
cl = predict_classes(res, k = 3, mat2, dist_method = "correlation")
#> The matrix has been scaled in cola analysis, thus the new matrix should
#> also be scaled with the same method ('z-score'). Please double check.
#> Set `help = FALSE` to suppress this message.
#>
#> * take top 500/2058 most significant signatures for prediction.
# compare the real classification and the predicted classification
data.frame(cola_class = get_classes(res, k = 3)[, "class"],
predicted = cl[, "class"])
#> cola_class predicted
#> 1 3 3
#> 2 3 3
#> 3 1 1
#> 4 1 1
#> 5 1 1
#> 6 3 3
#> 7 1 1
#> 8 1 1
#> 9 1 1
#> 10 1 1
#> 11 1 1
#> 12 1 1
#> 13 1 1
#> 14 1 1
#> 15 1 1
#> 16 1 1
#> 17 1 1
#> 18 3 3
#> 19 3 3
#> 20 1 1
#> 21 2 2
#> 22 2 2
#> 23 2 2
#> 24 2 2
#> 25 1 1
#> 26 2 2
#> 27 2 2
#> 28 1 1
#> 29 2 2
#> 30 2 2
#> 31 1 1
#> 32 2 2
#> 33 2 2
#> 34 2 2
#> 35 3 3
#> 36 1 1
#> 37 3 3
#> 38 3 3
#> 39 1 1
#> 40 3 3
#> 41 3 3
#> 42 3 3
#> 43 1 1
#> 44 1 1
#> 45 1 1
#> 46 2 2
#> 47 1 1
#> 48 1 1
#> 49 1 1
#> 50 1 1
#> 51 1 1
#> 52 3 3
#> 53 1 1
#> 54 1 1
#> 55 1 1
#> 56 2 2
#> 57 3 3
#> 58 1 1
#> 59 2 2
#> 60 1 1
#> 61 3 3
#> 62 2 2
#> 63 2 2
#> 64 2 2
#> 65 2 2
#> 66 2 2
#> 67 2 2
#> 68 1 1
#> 69 2 2
#> 70 2 2
#> 71 2 2
#> 72 2 2
# change to correlation method
cl = predict_classes(res, k = 3, mat2, dist_method = "correlation")
#> The matrix has been scaled in cola analysis, thus the new matrix should
#> also be scaled with the same method ('z-score'). Please double check.
#> Set `help = FALSE` to suppress this message.
#>
#> * take top 500/2058 most significant signatures for prediction.
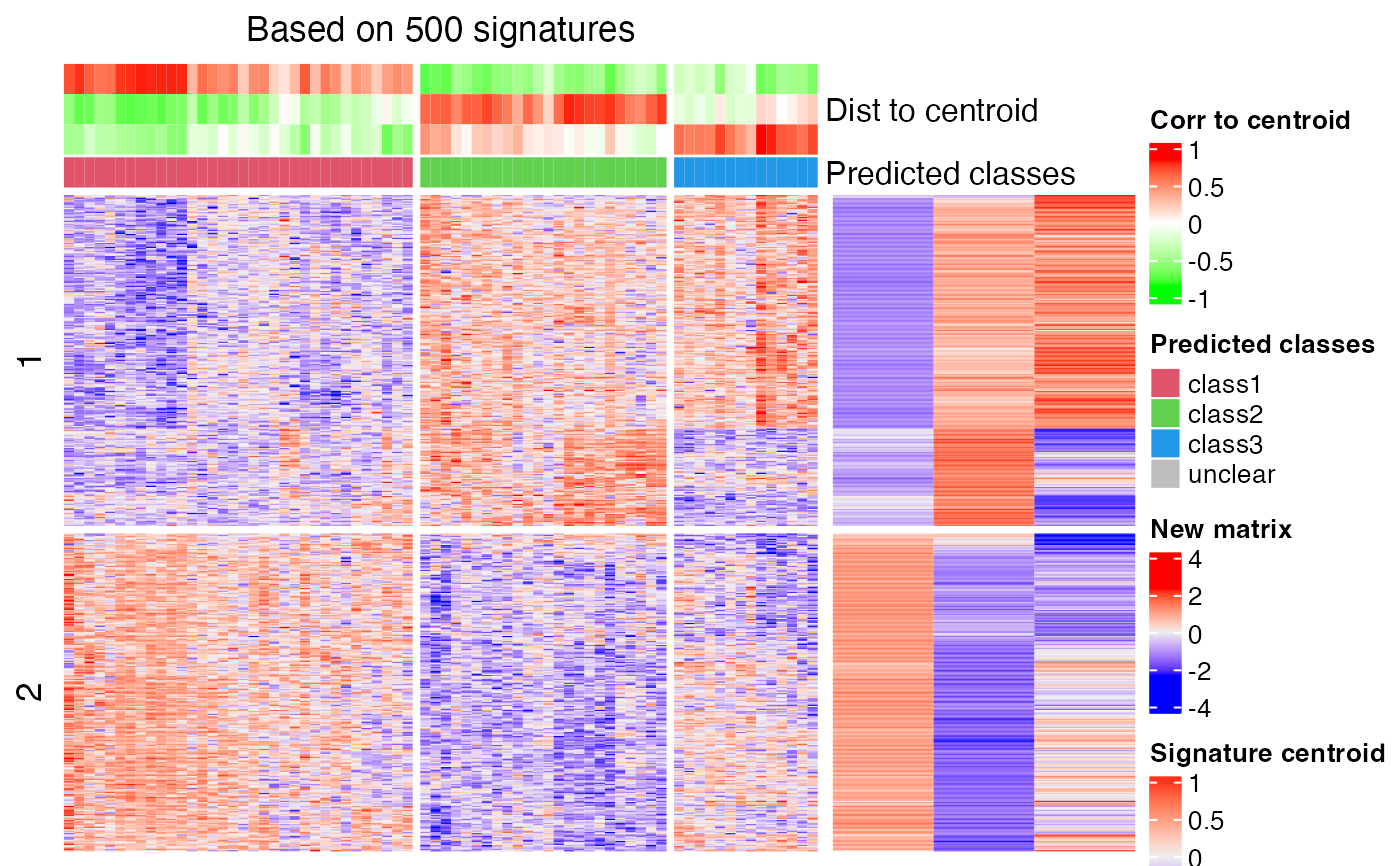 # compare the real classification and the predicted classification
data.frame(cola_class = get_classes(res, k = 3)[, "class"],
predicted = cl[, "class"])
#> cola_class predicted
#> 1 3 3
#> 2 3 3
#> 3 1 1
#> 4 1 1
#> 5 1 1
#> 6 3 3
#> 7 1 1
#> 8 1 1
#> 9 1 1
#> 10 1 1
#> 11 1 1
#> 12 1 1
#> 13 1 1
#> 14 1 1
#> 15 1 1
#> 16 1 1
#> 17 1 1
#> 18 3 3
#> 19 3 3
#> 20 1 1
#> 21 2 2
#> 22 2 2
#> 23 2 2
#> 24 2 2
#> 25 1 1
#> 26 2 2
#> 27 2 2
#> 28 1 1
#> 29 2 2
#> 30 2 2
#> 31 1 1
#> 32 2 2
#> 33 2 2
#> 34 2 2
#> 35 3 3
#> 36 1 1
#> 37 3 3
#> 38 3 3
#> 39 1 1
#> 40 3 3
#> 41 3 3
#> 42 3 3
#> 43 1 1
#> 44 1 1
#> 45 1 1
#> 46 2 2
#> 47 1 1
#> 48 1 1
#> 49 1 1
#> 50 1 1
#> 51 1 1
#> 52 3 3
#> 53 1 1
#> 54 1 1
#> 55 1 1
#> 56 2 2
#> 57 3 3
#> 58 1 1
#> 59 2 2
#> 60 1 1
#> 61 3 3
#> 62 2 2
#> 63 2 2
#> 64 2 2
#> 65 2 2
#> 66 2 2
#> 67 2 2
#> 68 1 1
#> 69 2 2
#> 70 2 2
#> 71 2 2
#> 72 2 2
# }
# compare the real classification and the predicted classification
data.frame(cola_class = get_classes(res, k = 3)[, "class"],
predicted = cl[, "class"])
#> cola_class predicted
#> 1 3 3
#> 2 3 3
#> 3 1 1
#> 4 1 1
#> 5 1 1
#> 6 3 3
#> 7 1 1
#> 8 1 1
#> 9 1 1
#> 10 1 1
#> 11 1 1
#> 12 1 1
#> 13 1 1
#> 14 1 1
#> 15 1 1
#> 16 1 1
#> 17 1 1
#> 18 3 3
#> 19 3 3
#> 20 1 1
#> 21 2 2
#> 22 2 2
#> 23 2 2
#> 24 2 2
#> 25 1 1
#> 26 2 2
#> 27 2 2
#> 28 1 1
#> 29 2 2
#> 30 2 2
#> 31 1 1
#> 32 2 2
#> 33 2 2
#> 34 2 2
#> 35 3 3
#> 36 1 1
#> 37 3 3
#> 38 3 3
#> 39 1 1
#> 40 3 3
#> 41 3 3
#> 42 3 3
#> 43 1 1
#> 44 1 1
#> 45 1 1
#> 46 2 2
#> 47 1 1
#> 48 1 1
#> 49 1 1
#> 50 1 1
#> 51 1 1
#> 52 3 3
#> 53 1 1
#> 54 1 1
#> 55 1 1
#> 56 2 2
#> 57 3 3
#> 58 1 1
#> 59 2 2
#> 60 1 1
#> 61 3 3
#> 62 2 2
#> 63 2 2
#> 64 2 2
#> 65 2 2
#> 66 2 2
#> 67 2 2
#> 68 1 1
#> 69 2 2
#> 70 2 2
#> 71 2 2
#> 72 2 2
# }