Compare clustering methods
Usage
compare_clustering_methods(
mat,
method = setdiff(all_clustering_methods(), "mclust"),
plot_type = c("mixed", "heatmap"),
nrow = 3,
verbose = TRUE
)Arguments
- mat
The similarity matrix.
- method
Which methods to compare. All available methods are in
all_clustering_methods. A value ofalltakes all available methods. By defaultmclustis excluded because its long runtime.- plot_type
See explanation in
cmp_make_plot.- nrow
Number of rows of the layout when
plot_typeis set toheatmap.- verbose
Whether to print messages.
Details
The function compares following clustering methods by default:
-kmeans see cluster_by_kmeans.
-pam see cluster_by_pam.
-dynamicTreeCut see cluster_by_dynamicTreeCut.
-mclust see cluster_by_mclust. By default it is not included.
-apcluster see cluster_by_apcluster.
-hdbscan see cluster_by_hdbscan.
-fast_greedy see cluster_by_fast_greedy.
-louvain see cluster_by_louvain.
-walktrap see cluster_by_walktrap.
-MCL see cluster_by_MCL.
-binary_cut see binary_cut.
This functon is basically a wrapper function. It calls the following two functions:
cmp_make_clusters: applies clustering with different methods.cmp_make_plot: makes the plots.
Examples
# \donttest{
mat = readRDS(system.file("extdata", "random_GO_BP_sim_mat.rds",
package = "simplifyEnrichment"))
compare_clustering_methods(mat)
#> Cluster 500 terms by 'kmeans'...
#> 18 clusters, used 3.221053 secs.
#> Cluster 500 terms by 'pam'...
#> 5 clusters, used 20.67218 secs.
#> Cluster 500 terms by 'dynamicTreeCut'...
#> 60 clusters, used 0.132031 secs.
#> Cluster 500 terms by 'apcluster'...
#> 41 clusters, used 0.7050171 secs.
#> Cluster 500 terms by 'hdbscan'...
#> 14 clusters, used 0.163321 secs.
#> Cluster 500 terms by 'fast_greedy'...
#> 6 clusters, used 0.06812 secs.
#> Cluster 500 terms by 'louvain'...
#> 6 clusters, used 0.07268381 secs.
#> Cluster 500 terms by 'walktrap'...
#> 6 clusters, used 0.780349 secs.
#> Cluster 500 terms by 'MCL'...
#> 6 clusters, used 1.736204 secs.
#> Cluster 500 terms by 'binary_cut'...
#> 19 clusters, used 4.215953 secs.
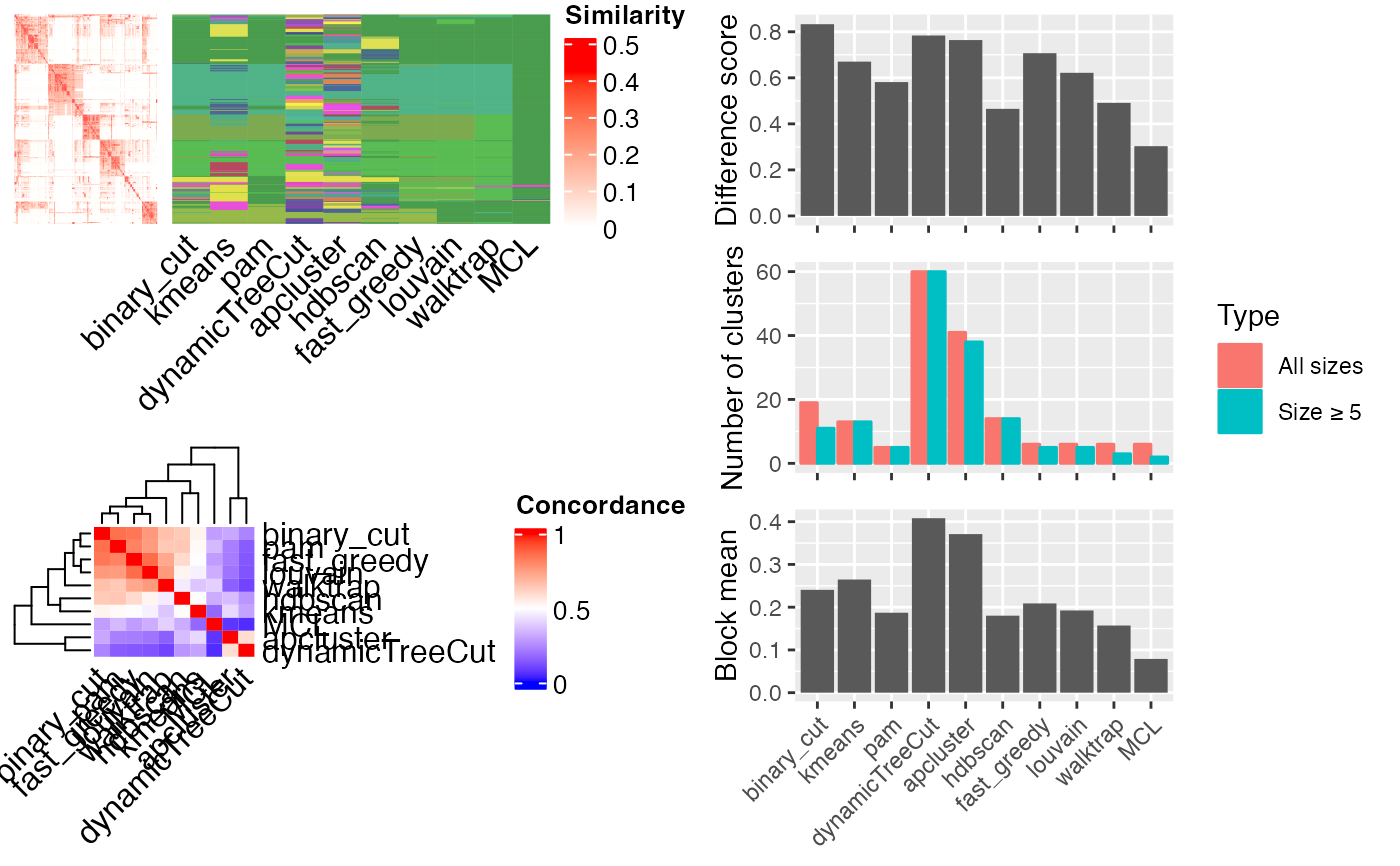 compare_clustering_methods(mat, plot_type = "heatmap")
#> Cluster 500 terms by 'kmeans'...
#> 23 clusters, used 3.011423 secs.
#> Cluster 500 terms by 'pam'...
#> 5 clusters, used 20.29424 secs.
#> Cluster 500 terms by 'dynamicTreeCut'...
#> 60 clusters, used 0.13887 secs.
#> Cluster 500 terms by 'apcluster'...
#> 41 clusters, used 0.7265048 secs.
#> Cluster 500 terms by 'hdbscan'...
#> 14 clusters, used 0.1619201 secs.
#> Cluster 500 terms by 'fast_greedy'...
#> 6 clusters, used 0.061584 secs.
#> Cluster 500 terms by 'louvain'...
#> 6 clusters, used 0.0815661 secs.
#> Cluster 500 terms by 'walktrap'...
#> 6 clusters, used 0.2488348 secs.
#> Cluster 500 terms by 'MCL'...
#> 6 clusters, used 1.787284 secs.
#> Cluster 500 terms by 'binary_cut'...
#> 19 clusters, used 5.14202 secs.
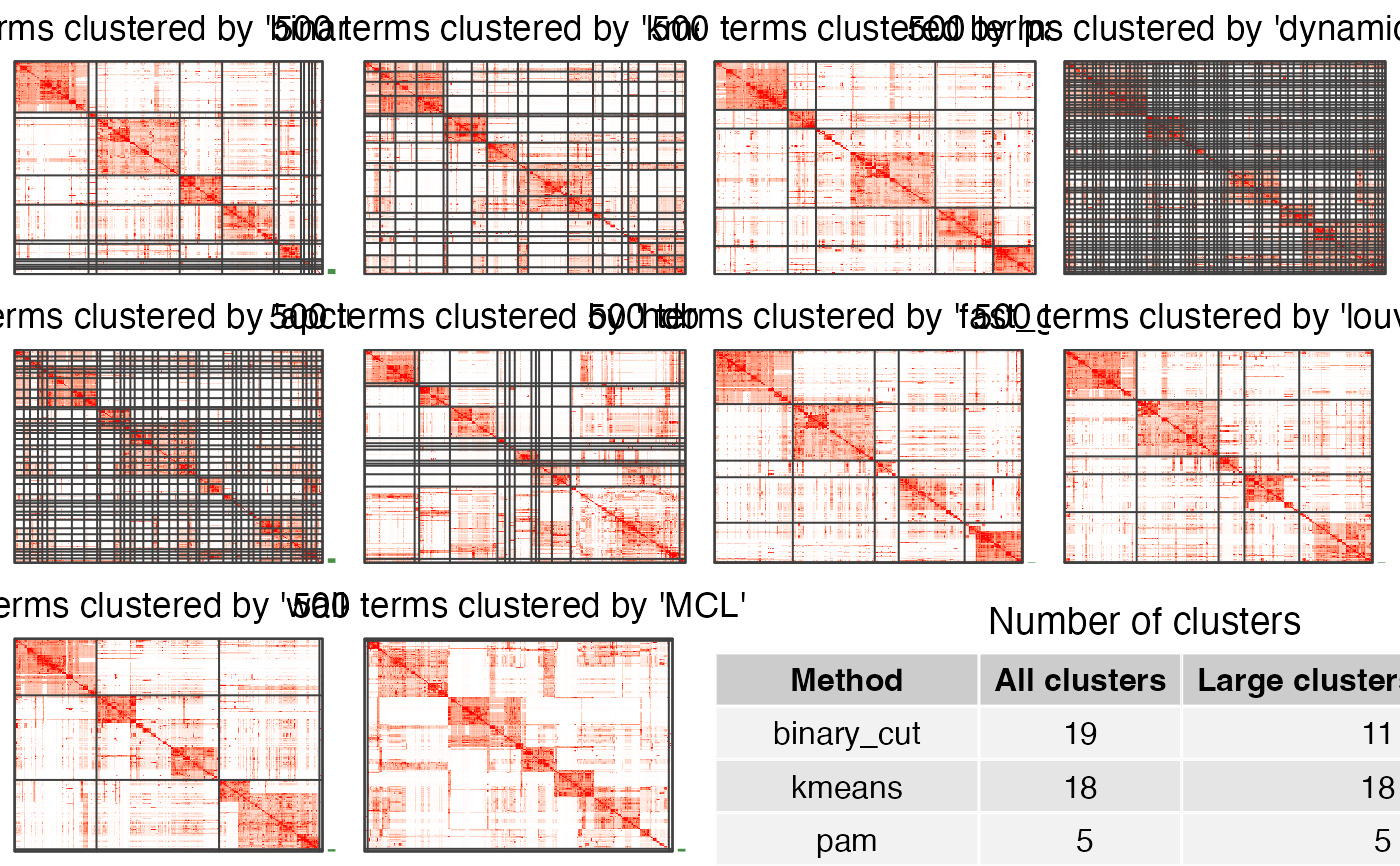
compare_clustering_methods(mat, plot_type = "heatmap")
#> Cluster 500 terms by 'kmeans'...
#> 23 clusters, used 3.011423 secs.
#> Cluster 500 terms by 'pam'...
#> 5 clusters, used 20.29424 secs.
#> Cluster 500 terms by 'dynamicTreeCut'...
#> 60 clusters, used 0.13887 secs.
#> Cluster 500 terms by 'apcluster'...
#> 41 clusters, used 0.7265048 secs.
#> Cluster 500 terms by 'hdbscan'...
#> 14 clusters, used 0.1619201 secs.
#> Cluster 500 terms by 'fast_greedy'...
#> 6 clusters, used 0.061584 secs.
#> Cluster 500 terms by 'louvain'...
#> 6 clusters, used 0.0815661 secs.
#> Cluster 500 terms by 'walktrap'...
#> 6 clusters, used 0.2488348 secs.
#> Cluster 500 terms by 'MCL'...
#> 6 clusters, used 1.787284 secs.
#> Cluster 500 terms by 'binary_cut'...
#> 19 clusters, used 5.14202 secs.
 # }
# }