Draw horizon chart along the spiral
spiral_horizon(
x,
y,
y_max = max(abs(y)),
n_slices = 4,
slice_size,
pos_fill = "#D73027",
neg_fill = "#313695",
use_bars = FALSE,
bar_width = min(diff(x)),
negative_from_top = FALSE,
track_index = current_track_index()
)Arguments
- x
X-locations of the data points.
- y
Y-locations of the data points.
- y_max
Maximal absolute value on y-axis.
- n_slices
Number of slices.
- slice_size
Size of the slices. The final number of sizes is
ceiling(max(abs(y))/slice_size).- pos_fill
Colors for positive values.
- neg_fill
Colors for negative values.
- use_bars
Whether to use bars?
- bar_width
Width of bars.
- negative_from_top
Should negative distribution be drawn from the top?
- track_index
Index of the track.
Value
A list of the following objects:
a color mapping function for colors.
a vector of intervals that split the data.
Details
Since the track height is very small in the spiral, horizon chart visualization is an efficient way to visualize distribution-like graphics.
See also
horizon_legend() for generating the legend.
Examples
# \donttest{
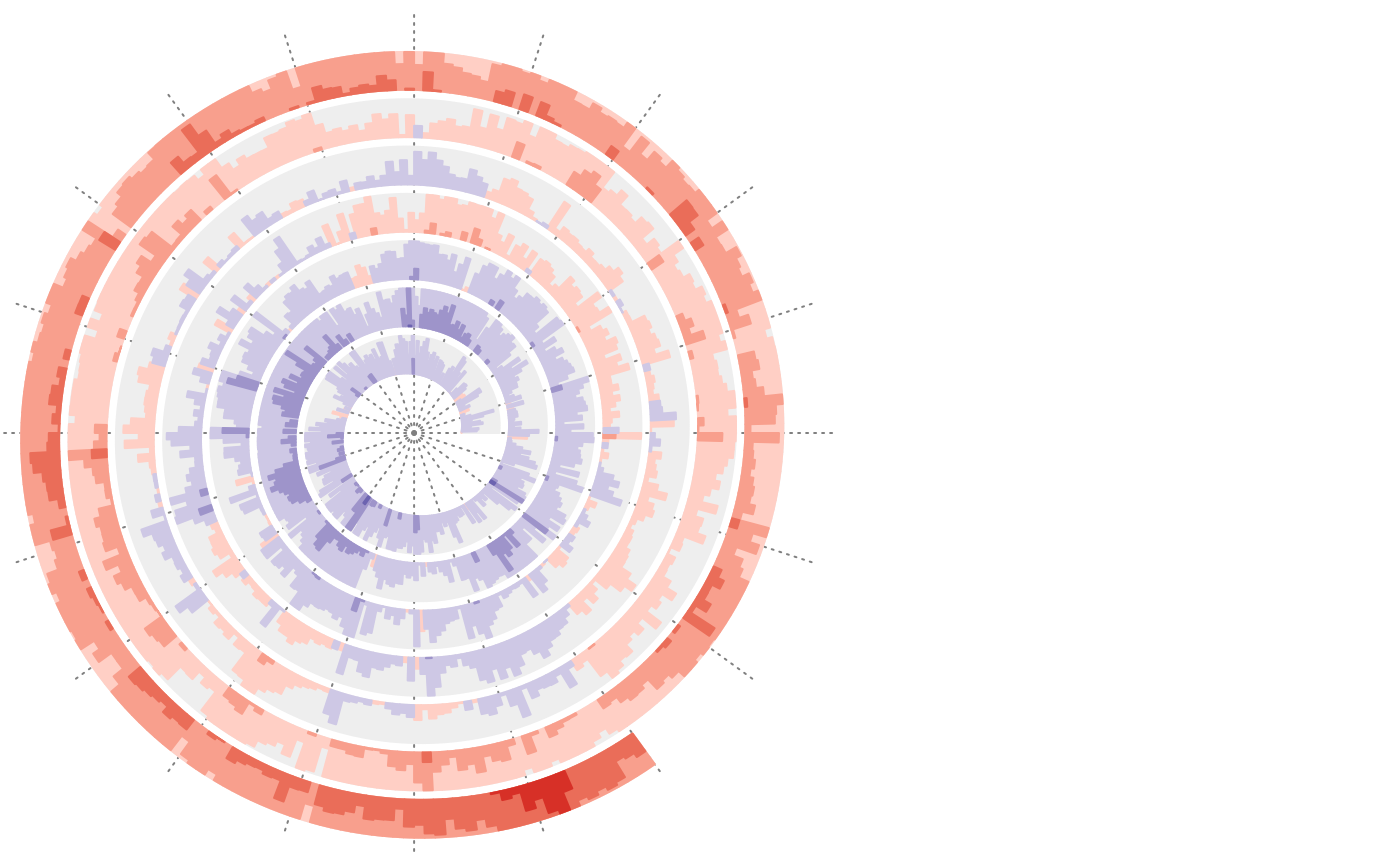
df = readRDS(system.file("extdata", "global_temperature.rds", package = "spiralize"))
df = df[df$Source == "GCAG", ]
spiral_initialize_by_time(xlim = range(df$Date), unit_on_axis = "months", period = "year",
period_per_loop = 20, polar_lines_by = 360/20)
spiral_track()
spiral_horizon(df$Date, df$Mean, use_bar = TRUE)
 # with legend
require(ComplexHeatmap)
#> Loading required package: ComplexHeatmap
#> ========================================
#> ComplexHeatmap version 2.18.0
#> Bioconductor page: http://bioconductor.org/packages/ComplexHeatmap/
#> Github page: https://github.com/jokergoo/ComplexHeatmap
#> Documentation: http://jokergoo.github.io/ComplexHeatmap-reference
#>
#> If you use it in published research, please cite either one:
#> - Gu, Z. Complex Heatmap Visualization. iMeta 2022.
#> - Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional
#> genomic data. Bioinformatics 2016.
#>
#>
#> The new InteractiveComplexHeatmap package can directly export static
#> complex heatmaps into an interactive Shiny app with zero effort. Have a try!
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(ComplexHeatmap))
#> ========================================
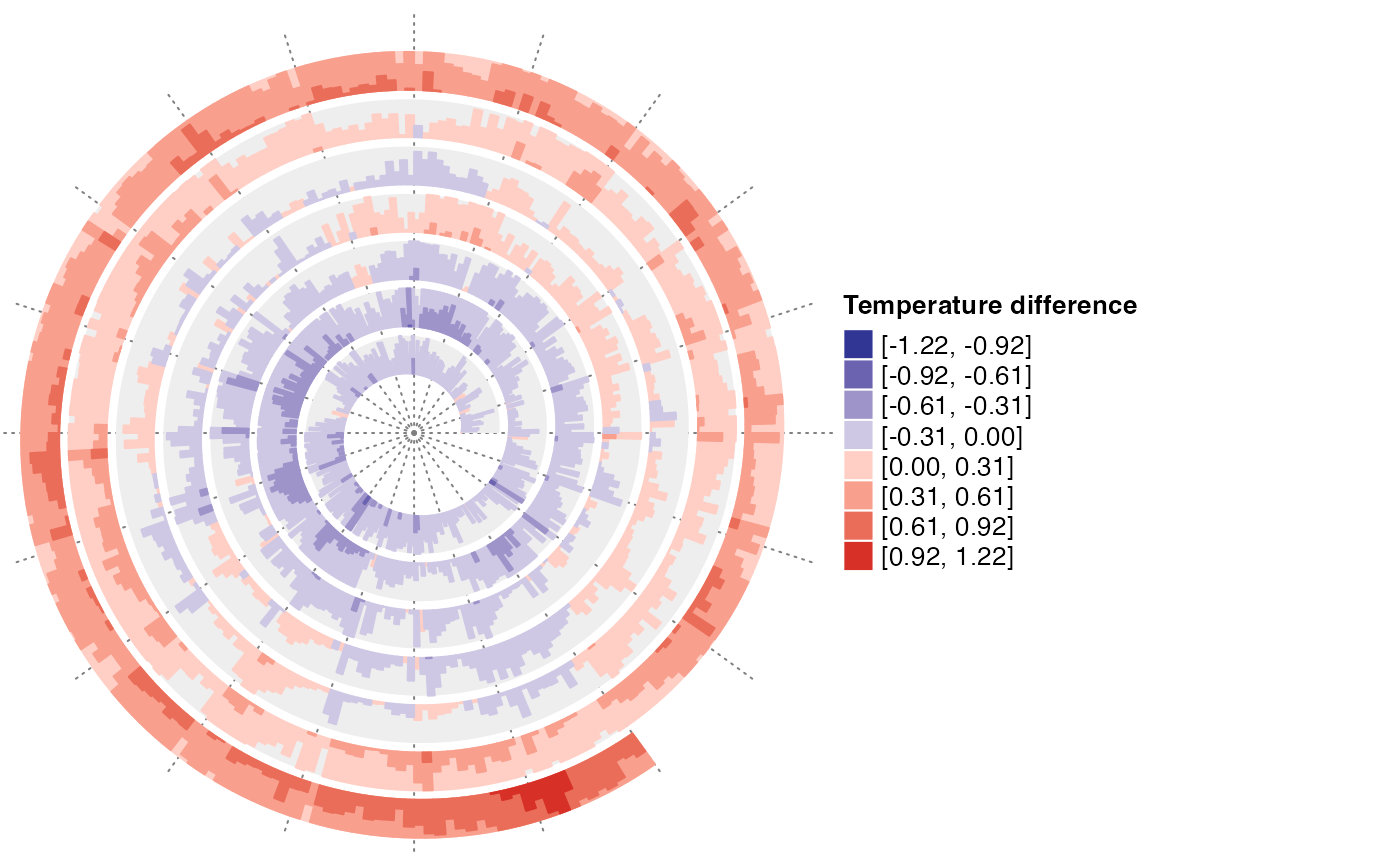
spiral_initialize_by_time(xlim = range(df$Date), unit_on_axis = "months", period = "year",
period_per_loop = 20, polar_lines_by = 360/20,
vp_param = list(x = unit(0, "npc"), just = "left"))
spiral_track()
lt = spiral_horizon(df$Date, df$Mean, use_bar = TRUE)
lgd = horizon_legend(lt, title = "Temperature difference")
draw(lgd, x = unit(1, "npc") + unit(2, "mm"), just = "left")
# with legend
require(ComplexHeatmap)
#> Loading required package: ComplexHeatmap
#> ========================================
#> ComplexHeatmap version 2.18.0
#> Bioconductor page: http://bioconductor.org/packages/ComplexHeatmap/
#> Github page: https://github.com/jokergoo/ComplexHeatmap
#> Documentation: http://jokergoo.github.io/ComplexHeatmap-reference
#>
#> If you use it in published research, please cite either one:
#> - Gu, Z. Complex Heatmap Visualization. iMeta 2022.
#> - Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional
#> genomic data. Bioinformatics 2016.
#>
#>
#> The new InteractiveComplexHeatmap package can directly export static
#> complex heatmaps into an interactive Shiny app with zero effort. Have a try!
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(ComplexHeatmap))
#> ========================================
spiral_initialize_by_time(xlim = range(df$Date), unit_on_axis = "months", period = "year",
period_per_loop = 20, polar_lines_by = 360/20,
vp_param = list(x = unit(0, "npc"), just = "left"))
spiral_track()
lt = spiral_horizon(df$Date, df$Mean, use_bar = TRUE)
lgd = horizon_legend(lt, title = "Temperature difference")
draw(lgd, x = unit(1, "npc") + unit(2, "mm"), just = "left")
 # }
# }