Initialize the spiral
spiral_initialize(
xlim = c(0, 1),
start = 360,
end = 360 * 5,
scale_by = c("angle", "curve_length"),
period = NULL,
clockwise = FALSE,
flip = c("none", "vertical", "horizontal", "both"),
reverse = FALSE,
polar_lines = scale_by == "angle",
polar_lines_by = 30,
polar_lines_gp = gpar(col = "#808080", lty = 3),
padding = unit(5, "mm"),
newpage = TRUE,
vp_param = list()
)Arguments
- xlim
Range on x-locations.
- start
Start of the spiral, in degree.
startandendshould be positive andstartshould be smaller thanend.- end
End of the spiral, in degree.
- scale_by
How scales on x-axis are equally interpolated? The values can be one of "angle" and "curve_length". If the value is "angle", equal angle difference corresponds to equal difference of data. In this case, in outer loops, the scales are longer than in the inner loops, although the difference on the data are the same. If the value is "curve_length", equal curve length difference corresponds to the equal difference of the data.
- period
Under "angle" mode, the number of loops can also be controlled by argument
periodwhich controls the length of data a spiral loop corresponds to. Note in this case, argumentendis ignored and the value forendis internally recalculated.- clockwise
Whether the curve is in a closewise direction. If it is set to
TRUE, argumentflipandreverseare ignored.- flip
How to flip the spiral? By default, the spiral starts from the origin of the coordinate and grows reverseclockwisely. The argument controls the growing direction of the spiral.
- reverse
By default, the most inside of the spiral corresponds to the lower boundary of x-location. Setting the value to
FALSEcan reverse the direction.- polar_lines
Whether draw the polar guiding lines.
- polar_lines_by
Increment of the polar lines. Measured in degree. The value can also be a vector that defines where to add polar lines.
- polar_lines_gp
Graphics parameters for the polar lines.
- padding
Padding of the plotting region. The value can be a
grid::unit()of length of one to two.- newpage
Whether to apply
grid::grid.newpage()before making the plot?- vp_param
A list of parameters sent to
grid::viewport().
Value
No value is returned.
Examples
spiral_initialize(); spiral_track()
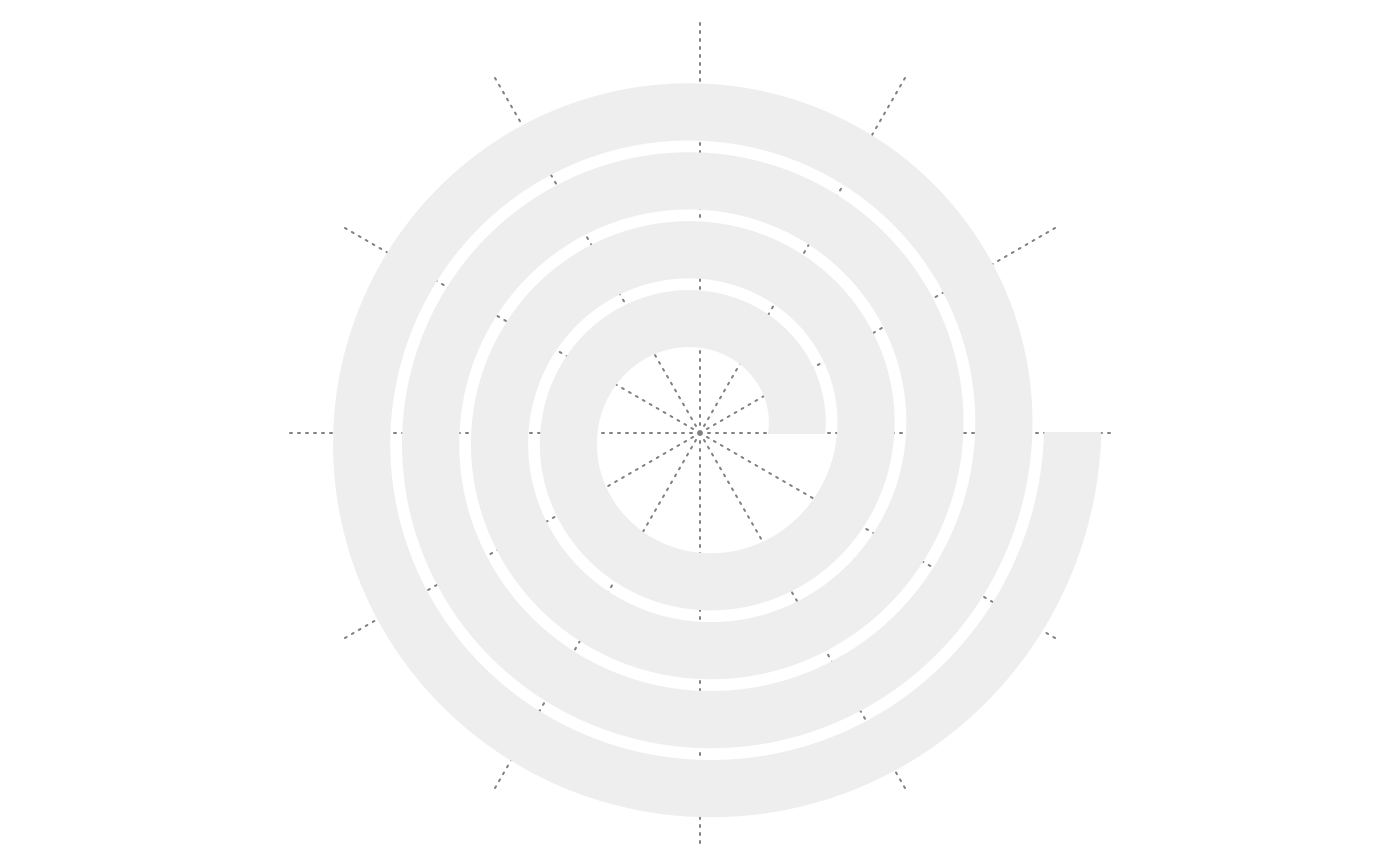 spiral_initialize(start = 180, end = 360+180); spiral_track()
spiral_initialize(start = 180, end = 360+180); spiral_track()
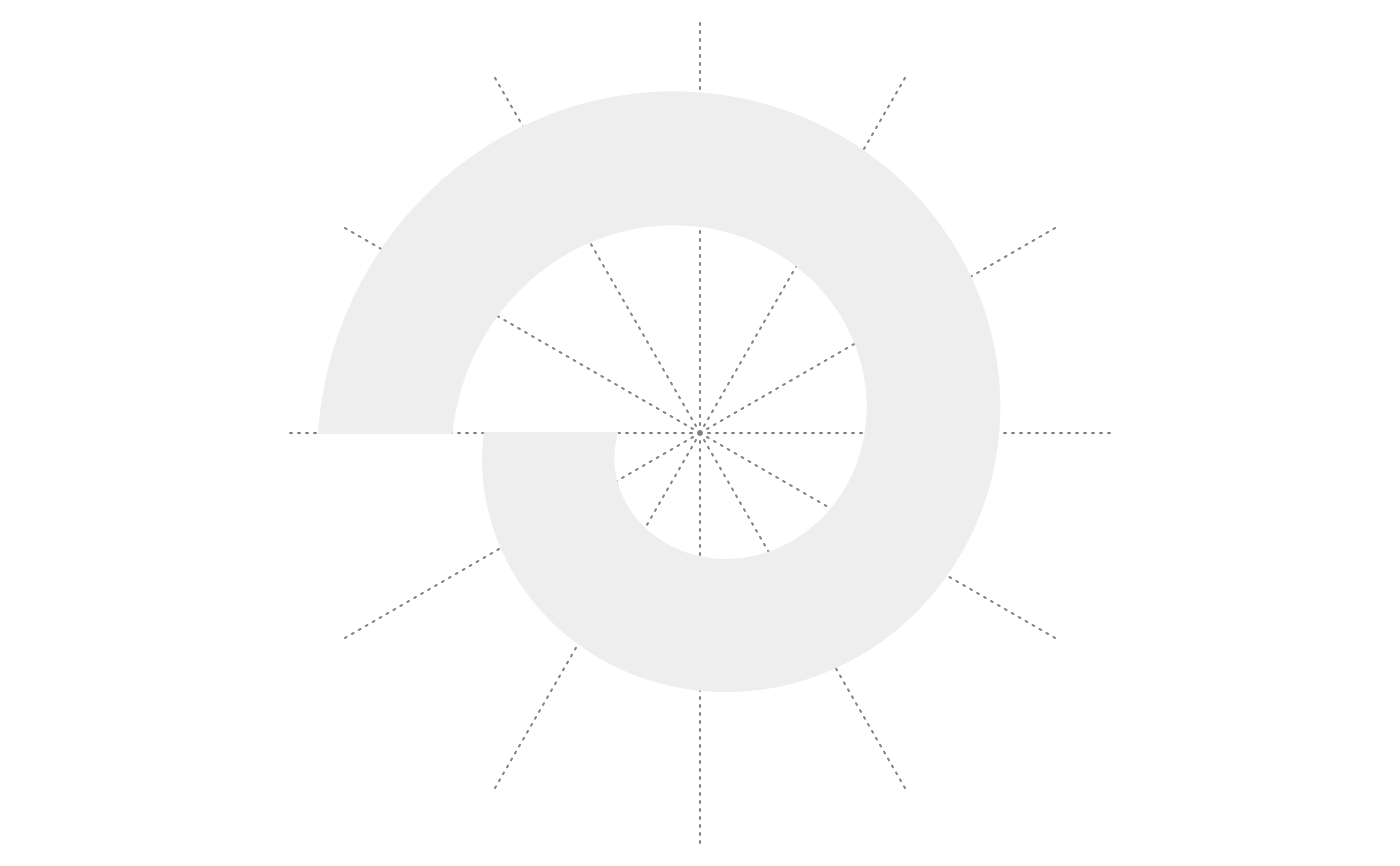 spiral_initialize(flip = "vertical"); spiral_track()
spiral_initialize(flip = "vertical"); spiral_track()
 spiral_initialize(flip = "horizontal"); spiral_track()
spiral_initialize(flip = "horizontal"); spiral_track()
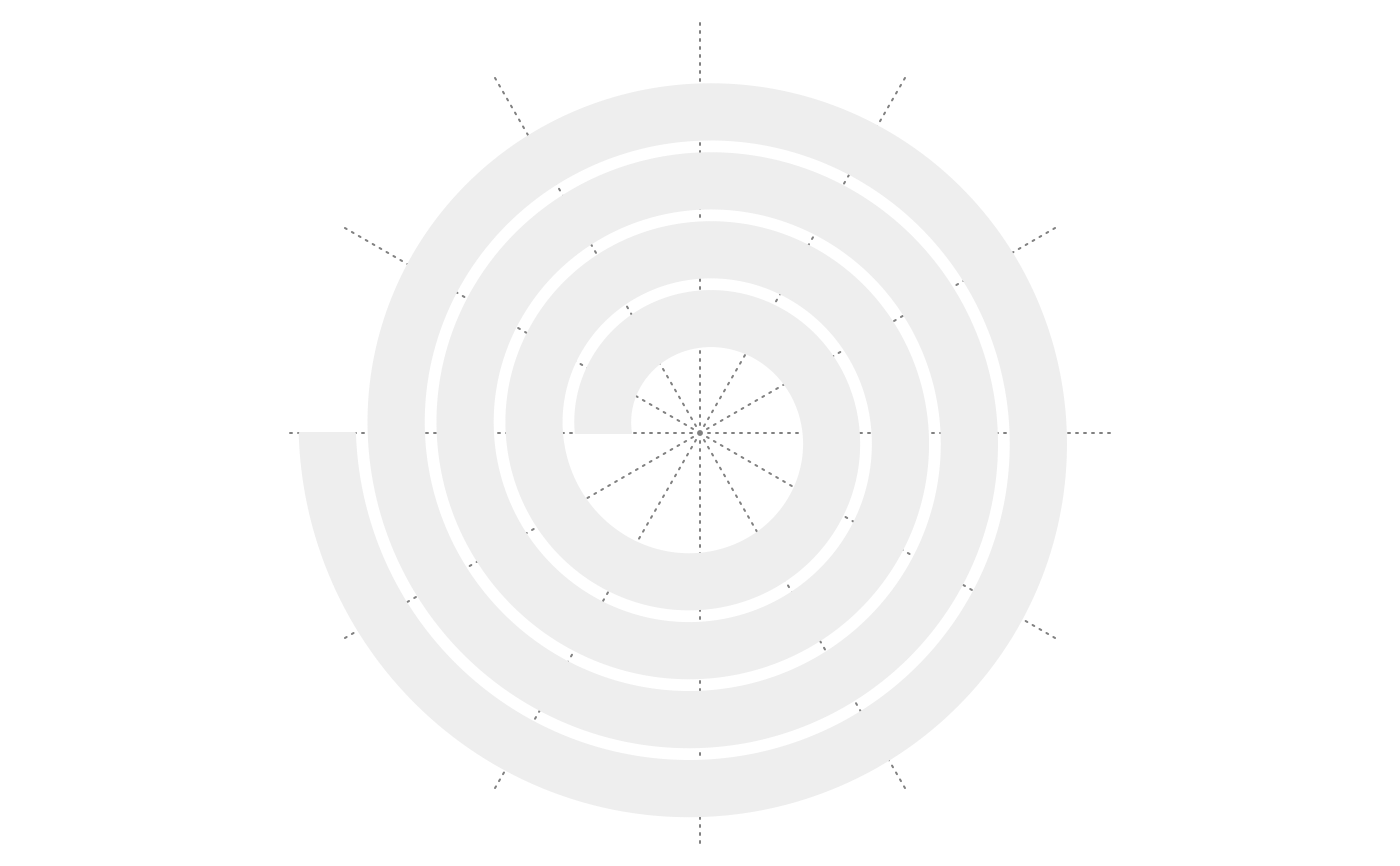 spiral_initialize(flip = "both"); spiral_track()
spiral_initialize(flip = "both"); spiral_track()
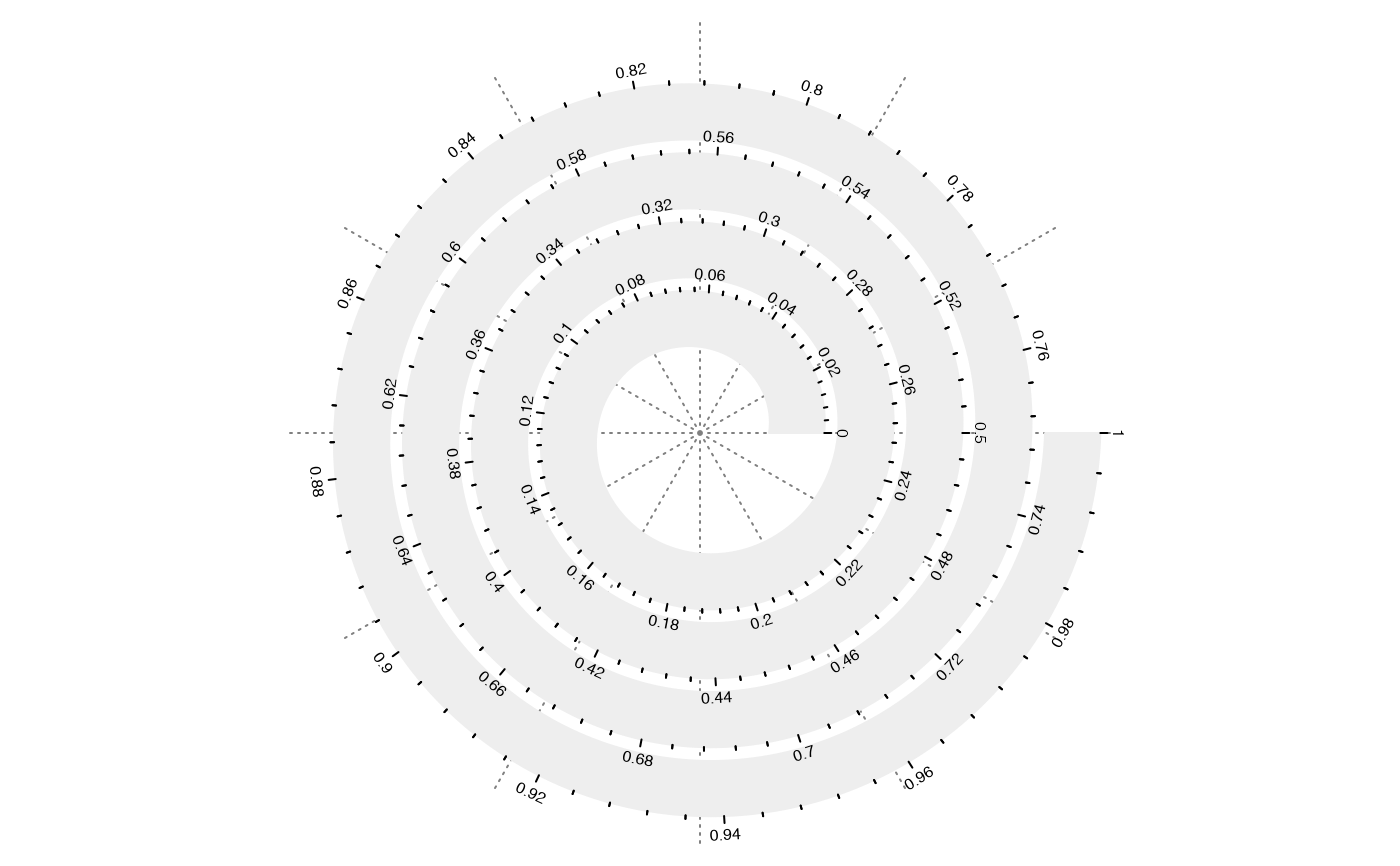
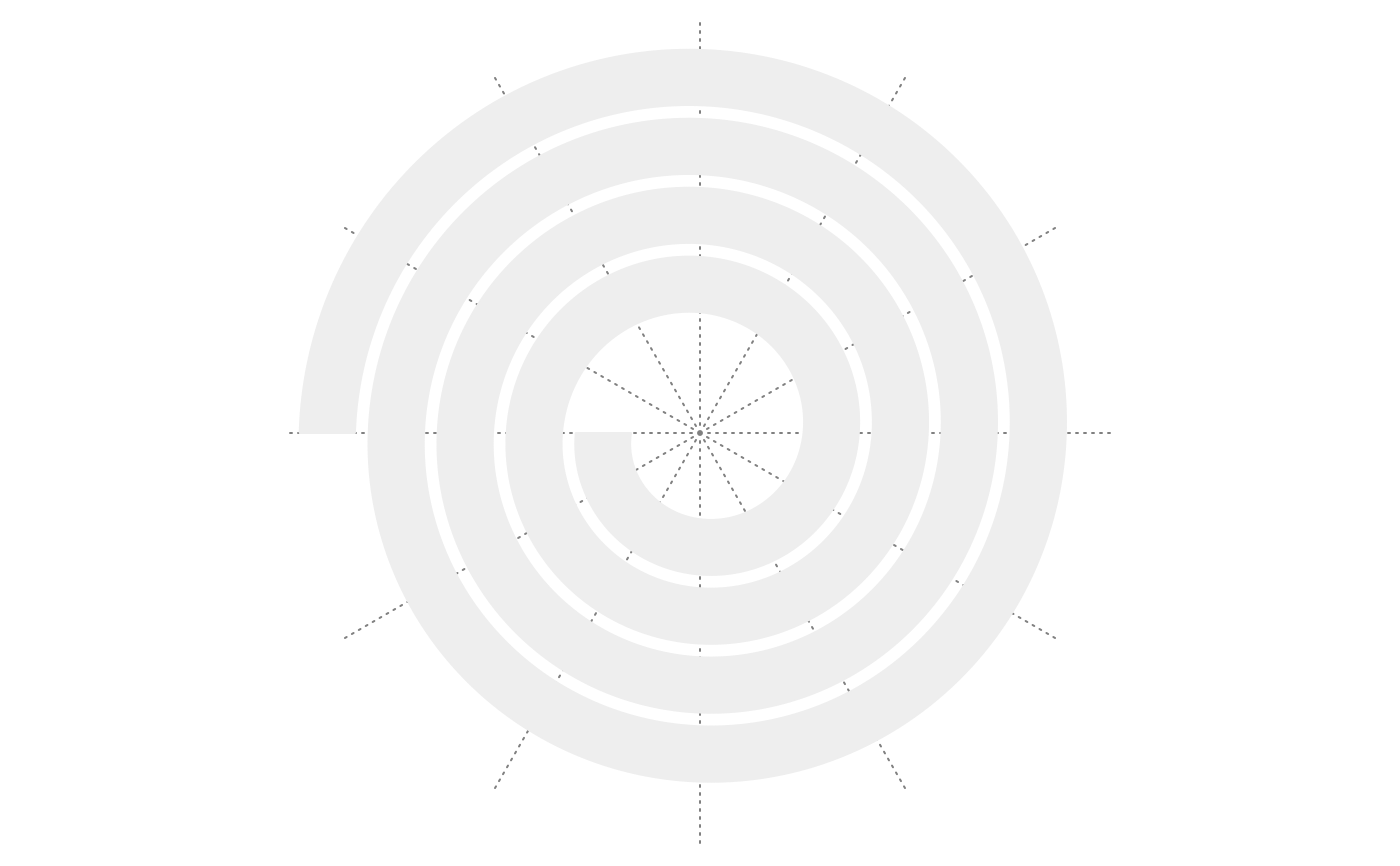 spiral_initialize(); spiral_track(); spiral_axis()
spiral_initialize(); spiral_track(); spiral_axis()
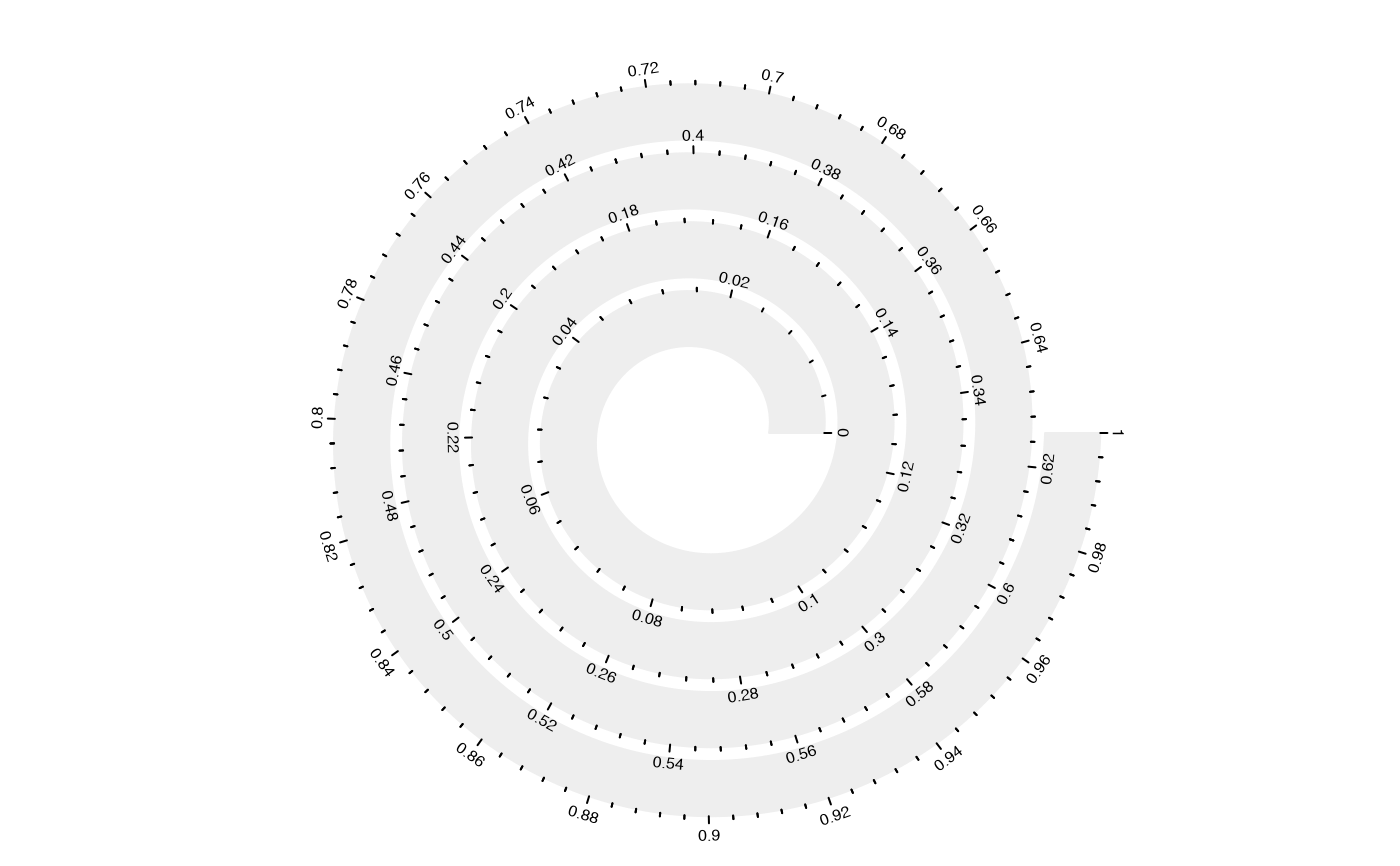
 spiral_initialize(scale_by = "curve_length"); spiral_track(); spiral_axis()
spiral_initialize(scale_by = "curve_length"); spiral_track(); spiral_axis()
 # the following example shows the difference of `scale_by` more clearly:
make_plot = function(scale_by) {
n = 100
require(circlize)
col = circlize::colorRamp2(c(0, 0.5, 1), c("blue", "white", "red"))
spiral_initialize(xlim = c(0, n), scale_by = scale_by)
spiral_track(height = 0.9)
x = runif(n)
spiral_rect(1:n - 1, 0, 1:n, 1, gp = gpar(fill = col(x), col = NA))
}
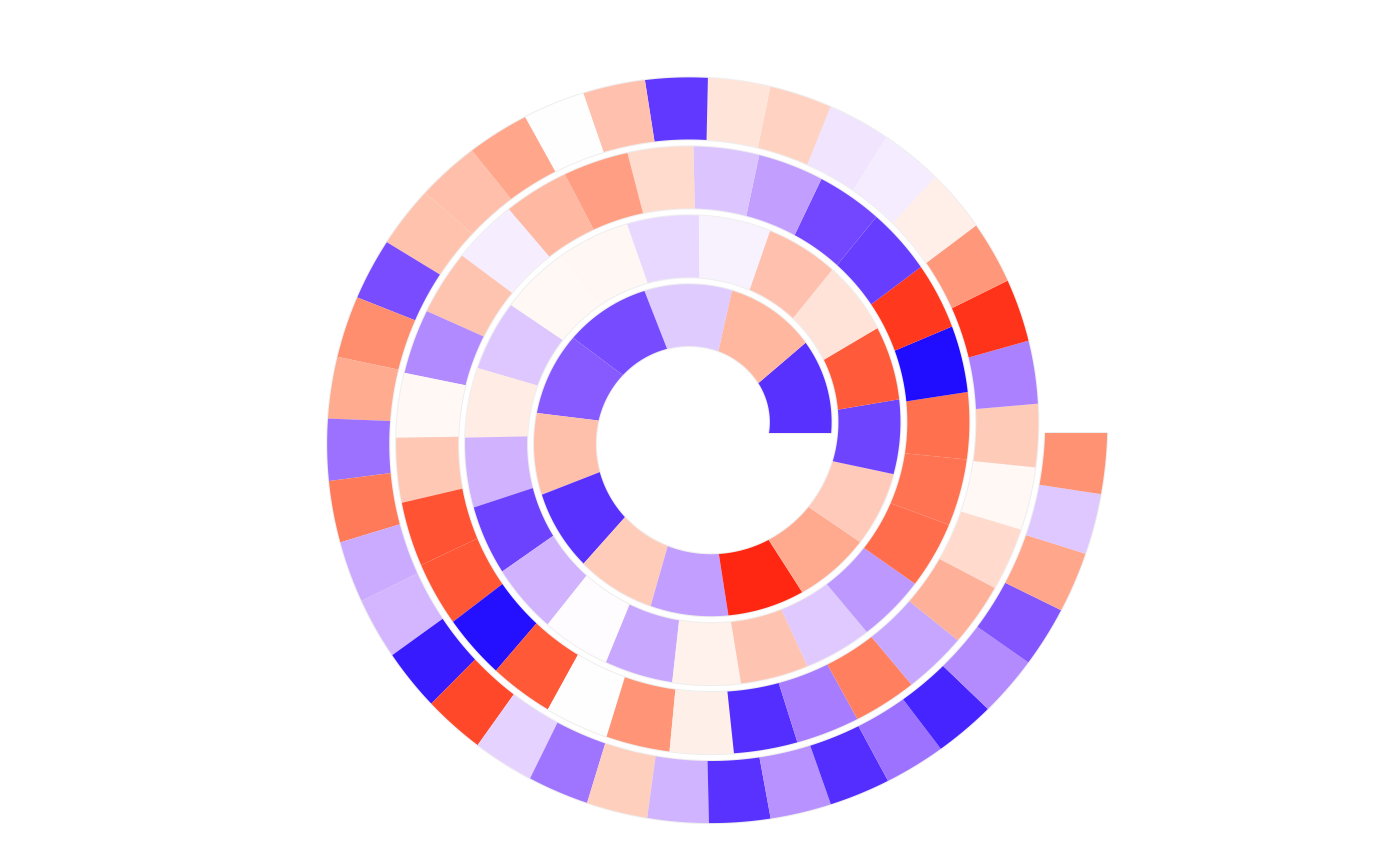
make_plot("angle")
#> Loading required package: circlize
#> ========================================
#> circlize version 0.4.16
#> CRAN page: https://cran.r-project.org/package=circlize
#> Github page: https://github.com/jokergoo/circlize
#> Documentation: https://jokergoo.github.io/circlize_book/book/
#>
#> If you use it in published research, please cite:
#> Gu, Z. circlize implements and enhances circular visualization
#> in R. Bioinformatics 2014.
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(circlize))
#> ========================================
# the following example shows the difference of `scale_by` more clearly:
make_plot = function(scale_by) {
n = 100
require(circlize)
col = circlize::colorRamp2(c(0, 0.5, 1), c("blue", "white", "red"))
spiral_initialize(xlim = c(0, n), scale_by = scale_by)
spiral_track(height = 0.9)
x = runif(n)
spiral_rect(1:n - 1, 0, 1:n, 1, gp = gpar(fill = col(x), col = NA))
}
make_plot("angle")
#> Loading required package: circlize
#> ========================================
#> circlize version 0.4.16
#> CRAN page: https://cran.r-project.org/package=circlize
#> Github page: https://github.com/jokergoo/circlize
#> Documentation: https://jokergoo.github.io/circlize_book/book/
#>
#> If you use it in published research, please cite:
#> Gu, Z. circlize implements and enhances circular visualization
#> in R. Bioinformatics 2014.
#>
#> This message can be suppressed by:
#> suppressPackageStartupMessages(library(circlize))
#> ========================================
 make_plot("curve_length")
make_plot("curve_length")