Draw phylogenetic tree
spiral_phylo(
obj,
gp = gpar(),
log = FALSE,
reverse = FALSE,
group = NULL,
group_col = NULL,
track_index = current_track_index()
)
phylo_to_dendrogram(obj, log = FALSE)Arguments
- obj
A
stats::dendrogramobject.- gp
Graphical parameters of the tree edges, mainly as a global setting.
- log
Whether the height of the tree to be log-transformed
log10(x + 1)?- reverse
Whether the tree to be reversed?
- group
A categorical variable for splitting the tree.
- group_col
A named vector which contains group colors.
- track_index
Index of the track.
Details
phylo_to_dendrogram() converts a phylo object to a dendrogram object.
The motivation is that phylogenetic tree may contain polytomies, which means at a certain node, there are more than two children branches. Available tools that do the conversion only support binary trees.
The returned dendrogram object is not in its standard format which means it can not be properly
drawn by the stats::plot.dendrogram() function. However, you can still apply stats::cutree() to the returned
dendrogram object with no problem and the dendrogram can be properly drawn with the ComplexHeatmap package (see examples).
Examples
require(ape)
#> Loading required package: ape
#>
#> Attaching package: ‘ape’
#> The following object is masked from ‘package:circlize’:
#>
#> degree
#> The following objects are masked from ‘package:dendextend’:
#>
#> ladderize, rotate
data(bird.families)
n = length(bird.families$tip.label)
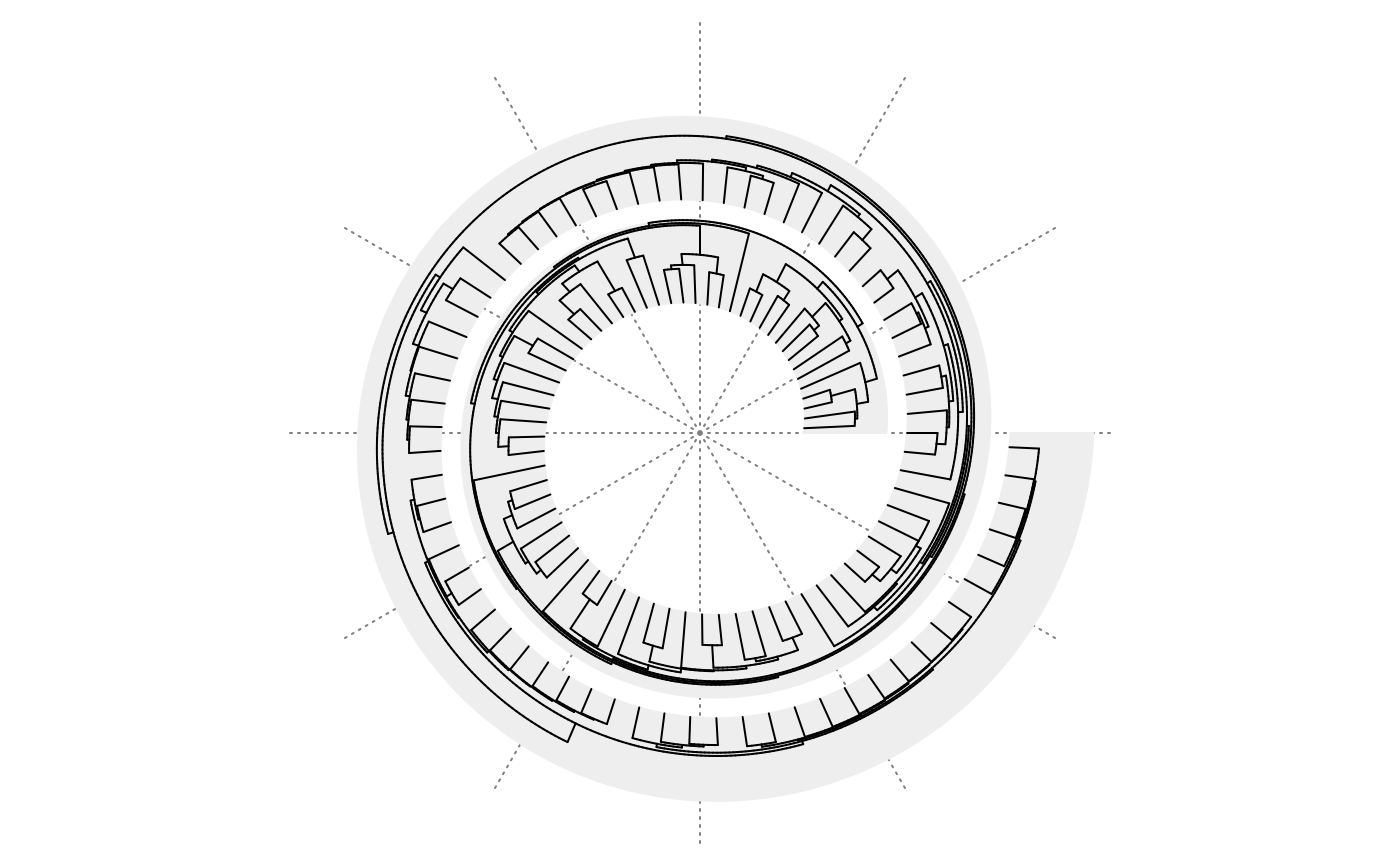
spiral_initialize(xlim = c(0, n), start = 360, end = 360*3)
spiral_track(height = 0.8)
spiral_phylo(bird.families)
 require(ape)
data(bird.families)
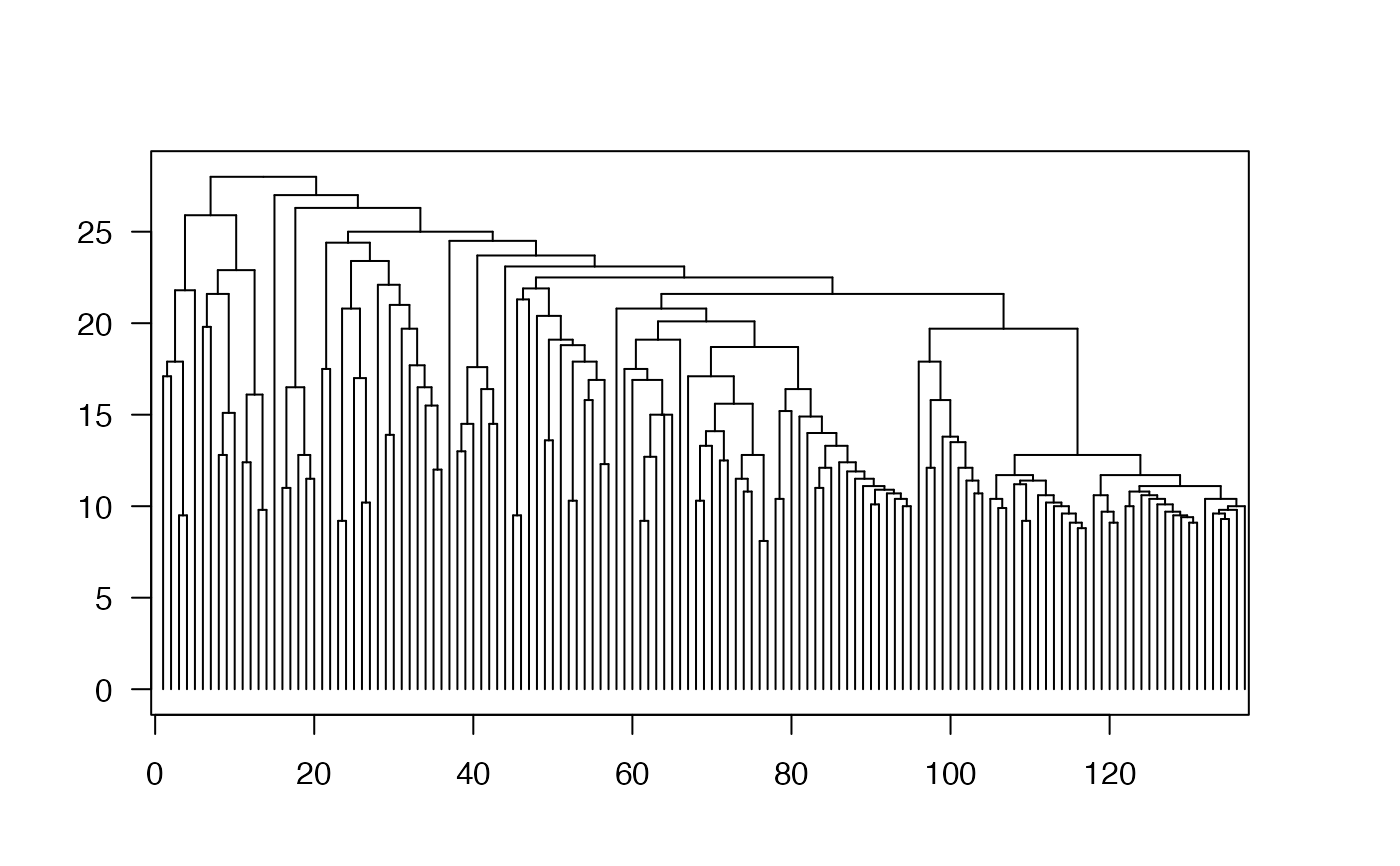
d = phylo_to_dendrogram(bird.families)
ComplexHeatmap::grid.dendrogram(d, test = TRUE)
require(ape)
data(bird.families)
d = phylo_to_dendrogram(bird.families)
ComplexHeatmap::grid.dendrogram(d, test = TRUE)