Topic 1-05: UniProt keywords gene sets
Zuguang Gu z.gu@dkfz.de
2025-05-31
Source:vignettes/topic1_05_UniProtKeywords.Rmd
topic1_05_UniProtKeywords.RmdUniProt database provides a list of controlled vocabulary represented as keywords for genes or proteins (https://www.uniprot.org/keywords/). This is useful for summarizing gene functions in a compact way.
The UniProtKeywords package
First load the package:
The release and source information of the data:
UniProtKeywords## UniProt Keywords
## Release: 2023_04 of 13-Sep-2023
## Source: https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/docs/keywlist.txt
## Number of keywords: 1201
## Built date: 2023-09-23The UniProtKeywords package has compiled genesets of
keywords for some organism, which can get by the function
load_keyword_genesets(). The argument is the taxon ID of a
organism. The full set of supported organisms can be found in the
document of load_keyword_genesets() (or in the object
UniProtKeywords:::ORGANISM).
gl = load_keyword_genesets(9606)
gl[sample(length(gl), 2)]## $`Heparin-binding`
## [1] "351" "51816" "9289" "4043" "27329" "462" "26" "333"
## [9] "338" "348" "350" "339366" "9510" "9508" "566" "6359"
## [17] "6368" "6354" "6355" "3491" "1490" "1289" "1305" "1311"
## [25] "84570" "6372" "2160" "129804" "2200" "2246" "2247" "2249"
## [33] "2252" "2254" "143282" "2260" "2263" "2335" "11167" "5045"
## [41] "80739" "2660" "5270" "64388" "1839" "3053" "3273" "3617"
## [49] "50939" "5104" "3730" "3918" "3990" "9388" "200879" "149998"
## [57] "4023" "4053" "56925" "4192" "8829" "8828" "26577" "5197"
## [65] "5196" "5228" "10631" "5549" "5553" "400668" "5764" "5792"
## [73] "5802" "6146" "6159" "284654" "340419" "84870" "343637" "6288"
## [81] "50964" "4057" "7057" "7058" "7422" "7423" "7448" "51156"
##
## $`rRNA-binding`
## [1] "284119" "55794" "26284" "22868" "60493" "55272" "84678" "51776"
## [9] "10436" "79159" "387338" "55037" "81554" "6135" "6147" "6167"
## [17] "6125" "6132" "80135" "6205" "6222" "6191" "6192" "140032"
## [25] "6203" "51373" "23107"You can also use the name of the organism:
load_keyword_genesets("human")
load_keyword_genesets("Homo sapiens")Argument as_table can be set to TRUE, then
load_keyword_genesets() returns a two-column data
frame.
tb = load_keyword_genesets(9606, as_table = TRUE)
head(tb)## keyword gene
## 1 2Fe-2S 2230
## 2 2Fe-2S 316
## 3 2Fe-2S 55847
## 4 2Fe-2S 493856
## 5 2Fe-2S 284106
## 6 2Fe-2S 57019Statistics
We can simply check some statistics.
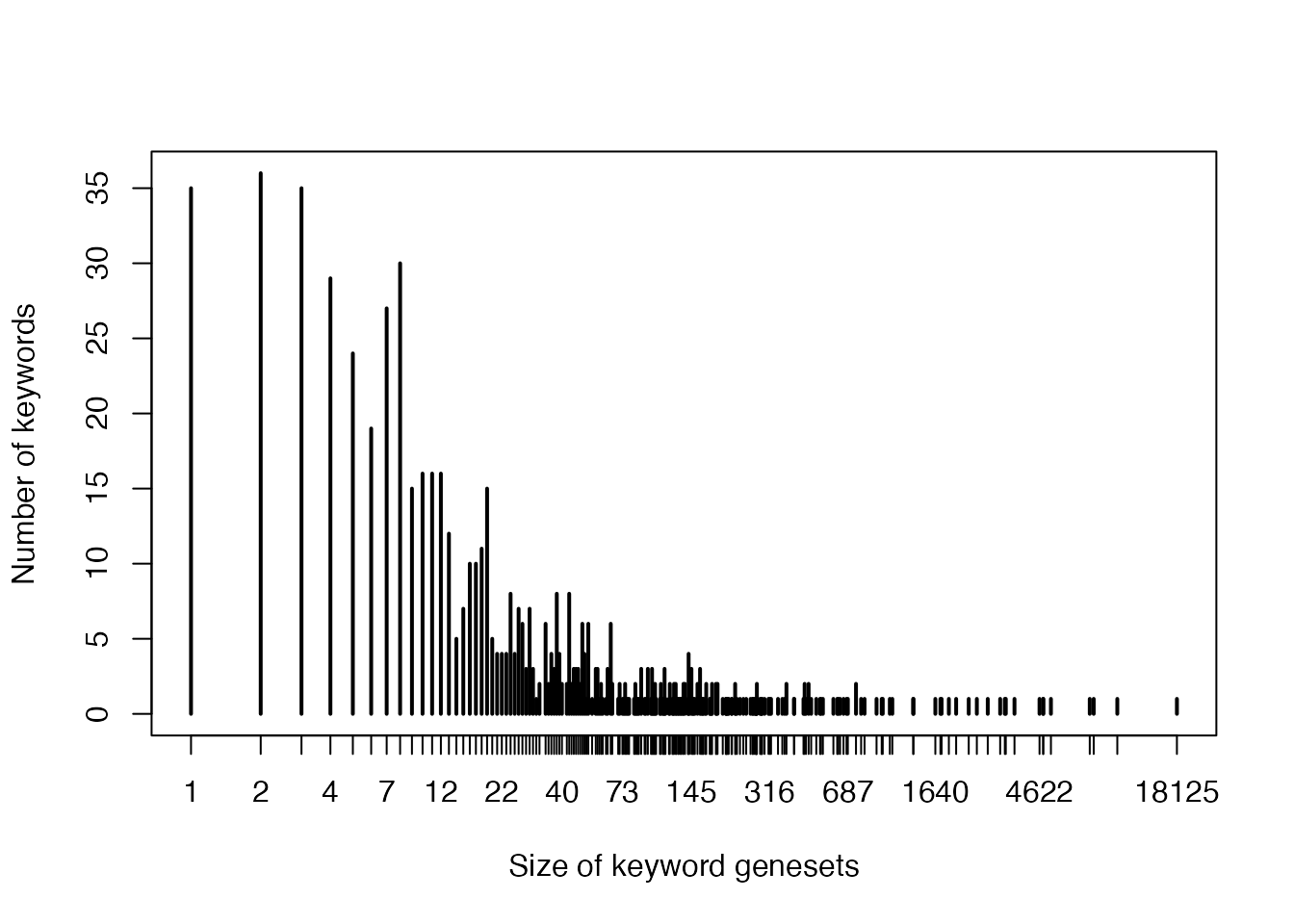
- Sizes of keyword genesets:
plot(table(sapply(gl, length)), log = "x",
xlab = "Size of keyword genesets",
ylab = "Number of keywords"
)
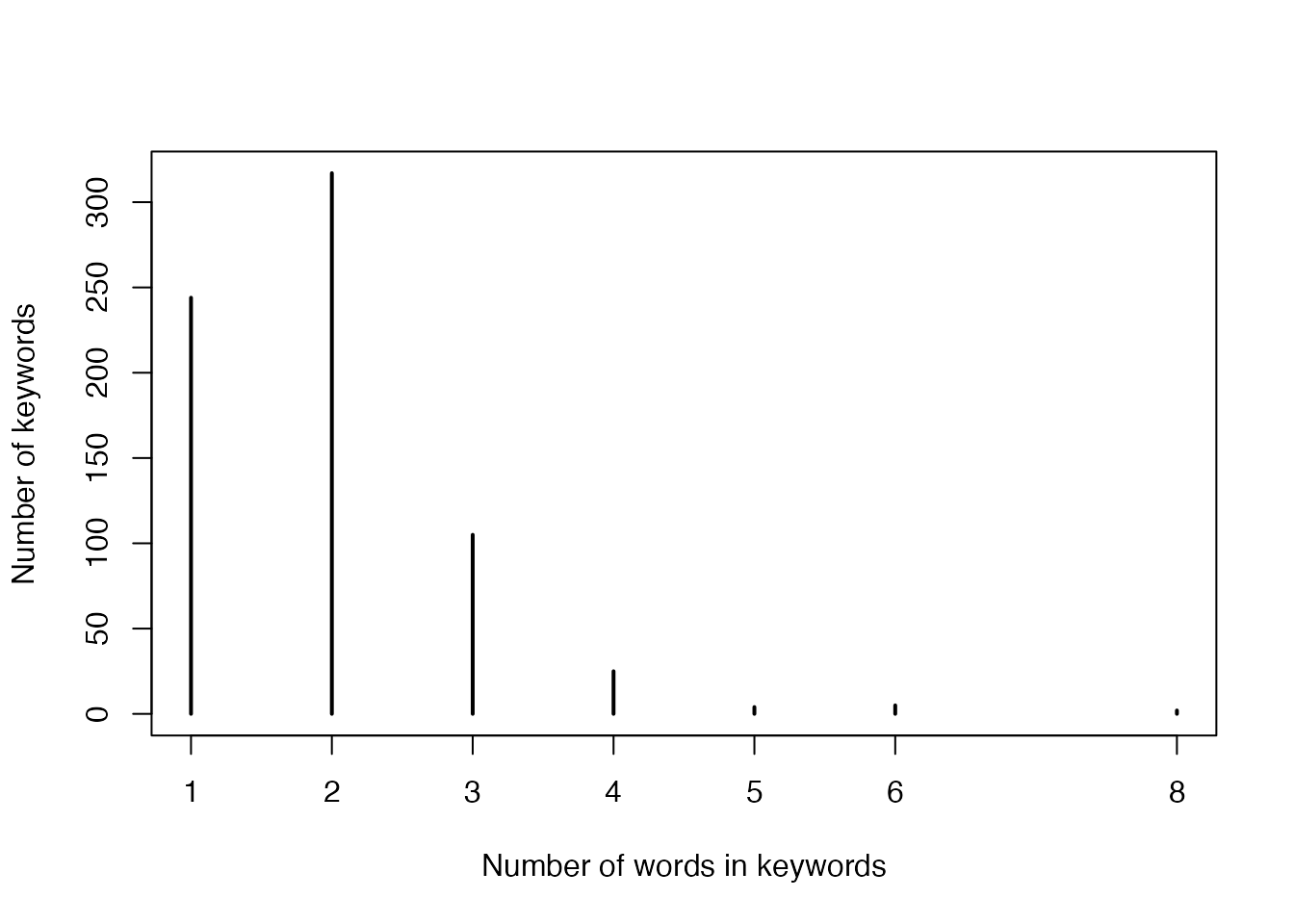
- Numbers of words in keywords:
plot(table(sapply(strsplit(names(gl), " |-|/"), length)),
xlab = "Number of words in keywords",
ylab = "Number of keywords"
)
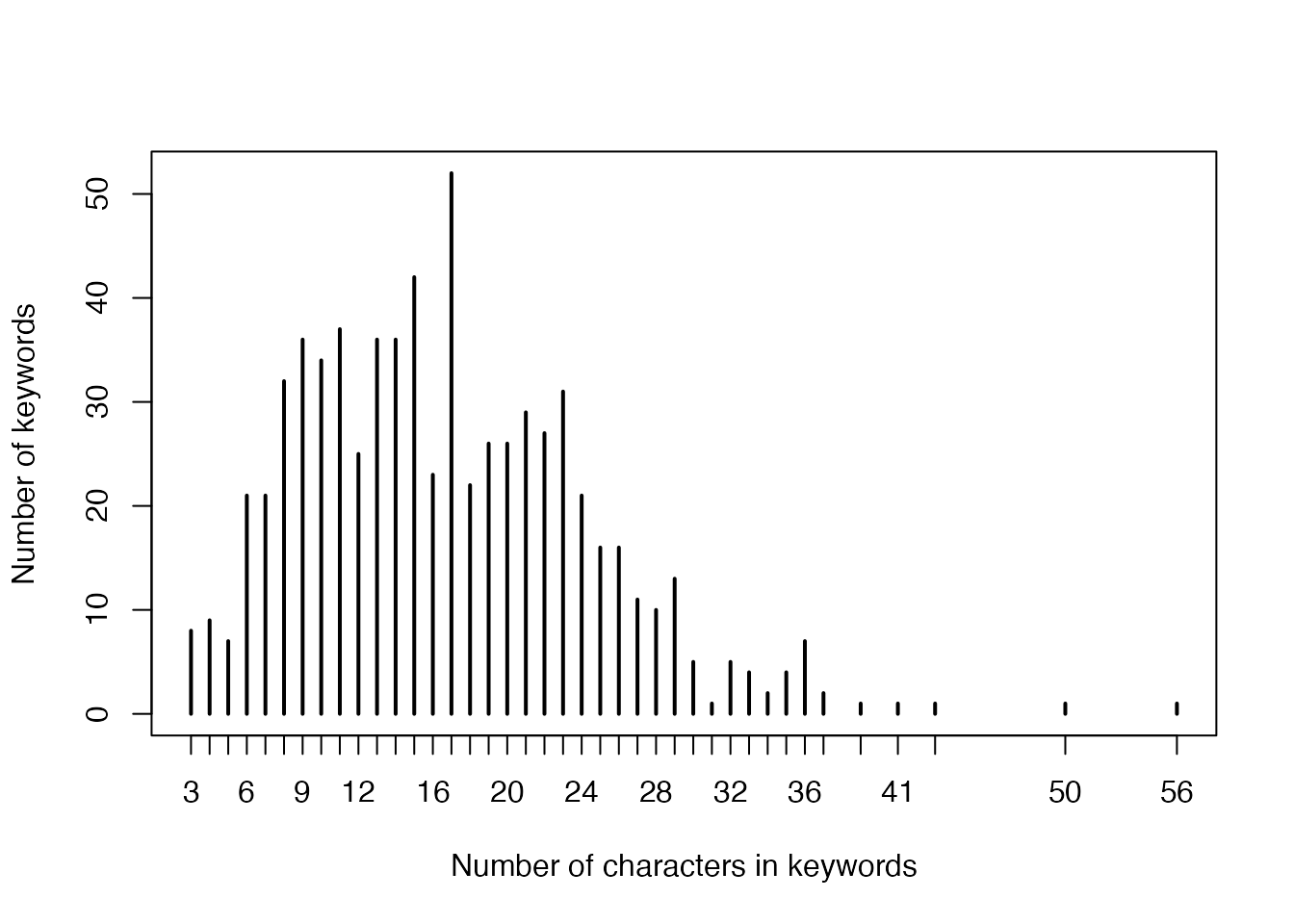
- Numbers of characters in keywords:
plot(table(nchar(names(gl))),
xlab = "Number of characters in keywords",
ylab = "Number of keywords"
)
Practice
Practice 1
What are the keywords with more than 2000 genes or 35 characters?
len = sapply(gl, length)
len[len > 2000]## 3D-structure Acetylation Alternative splicing
## 7617 3305 10018
## Coiled coil Direct protein sequencing Disease variant
## 2017 2762 3603
## Disulfide bond Membrane Phosphoprotein
## 3273 5177 7922
## Reference proteome Repeat Signal
## 18125 4622 3125
## Transcription Transmembrane Transmembrane helix
## 2286 4797 4793
## Ubl conjugation
## 2478## [1] "Activation of host autophagy by virus"
## [2] "Branched-chain amino acid catabolism"
## [3] "Complement activation lectin pathway"
## [4] "Congenital disorder of glycosylation"
## [5] "Congenital generalized lipodystrophy"
## [6] "Congenital stationary night blindness"
## [7] "Familial hemophagocytic lymphohistiocytosis"
## [8] "Hereditary nonpolyposis colorectal cancer"
## [9] "Inhibition of host innate immune response by virus"
## [10] "Inhibition of host interferon signaling pathway by virus"
## [11] "Lacrimo-auriculo-dento-digital syndrome"
## [12] "Microtubular inwards viral transport"
## [13] "Progressive external ophthalmoplegia"
## [14] "Rhizomelic chondrodysplasia punctata"