Topic 1-06: Get gene sets from UniProt Keywords
Zuguang Gu z.gu@dkfz.de
2025-03-02
Source:vignettes/topic1_06_UniProtKeywords.Rmd
topic1_06_UniProtKeywords.RmdUniProt database provides a list of controlled vocabulary represented as keywords for genes or proteins (https://www.uniprot.org/keywords/). This is useful for summarizing gene functions in a compact way.
The UniProtKeywords package
First load the package:
The release and source information of the data:
UniProtKeywords## UniProt Keywords
## Release: 2023_04 of 13-Sep-2023
## Source: https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/docs/keywlist.txt
## Number of keywords: 1201
## Built date: 2023-09-23The UniProtKeywords package has compiled genesets of keywords for some organism, which can get by the function load_keyword_genesets(). The argument is the taxon ID of a organism. The full set of supported organisms can be found in the document of load_keyword_genesets() (or in the object UniProtKeywords:::ORGANISM).
gl = load_keyword_genesets(9606)
gl[3:4] # because gl[1:2] has a very long output, here we print gl[3:4]## $`3Fe-4S`
## [1] "6390"
##
## $`4Fe-4S`
## [1] "6059" "48" "50" "54901" "64428" "51654" "57019" "1663"
## [9] "1763" "5424" "5426" "1806" "55140" "2068" "2110" "64789"
## [17] "83990" "3658" "11019" "4337" "4595" "4719" "4720" "374291"
## [25] "4728" "4723" "4682" "10101" "5558" "5471" "55316" "91543"
## [33] "51750" "6390" "441250" "55253"You can also use the name of the organism:
load_keyword_genesets("human")
load_keyword_genesets("Homo sapiens")Argument as_table can be set to TRUE, then load_keyword_genesets() returns a two-column data frame.
tb = load_keyword_genesets(9606, as_table = TRUE)
head(tb)## keyword gene
## 1 2Fe-2S 2230
## 2 2Fe-2S 316
## 3 2Fe-2S 55847
## 4 2Fe-2S 493856
## 5 2Fe-2S 284106
## 6 2Fe-2S 57019Statistics
We can simply check some statistics.
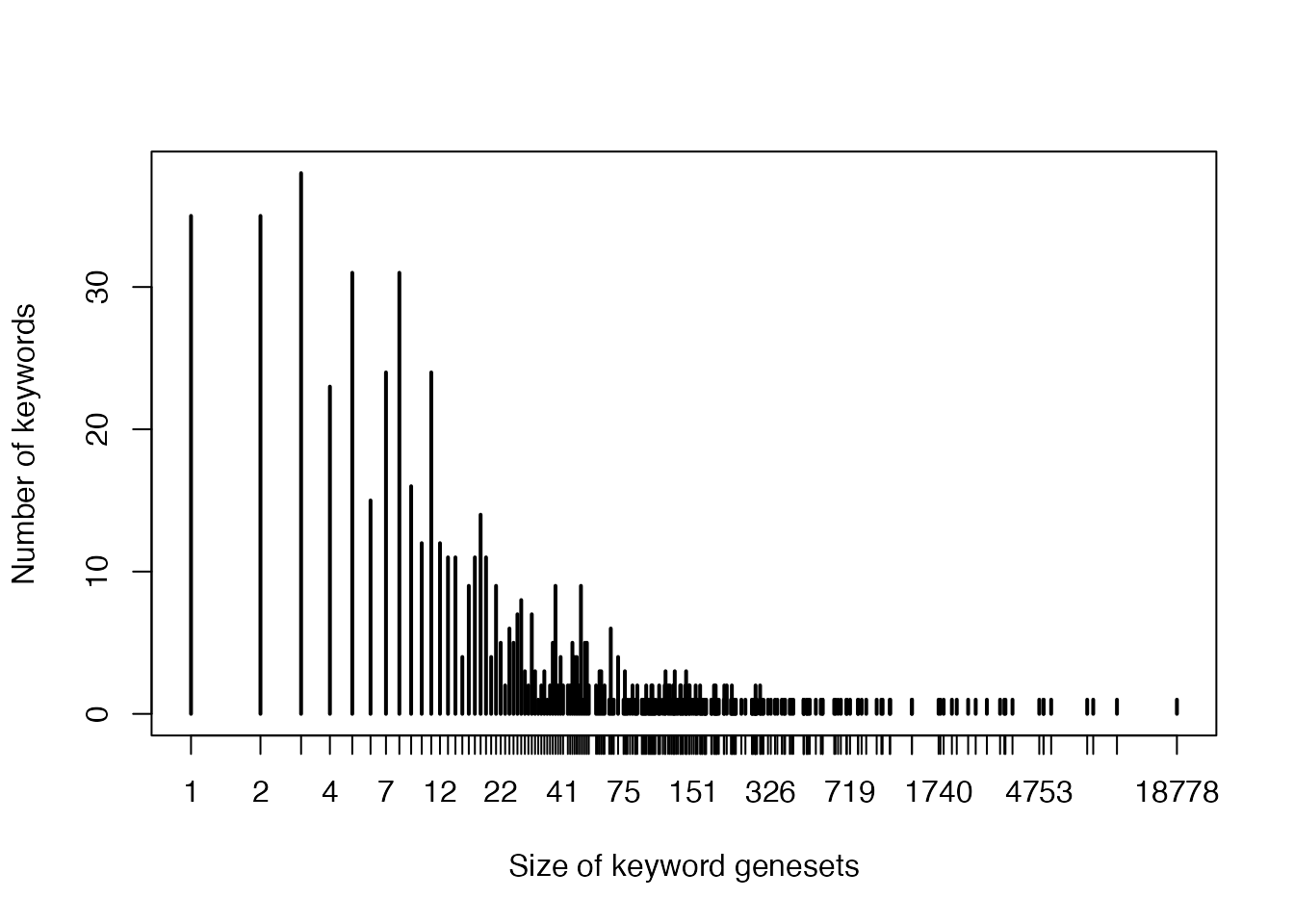
- Sizes of keyword genesets:
plot(table(sapply(gl, length)), log = "x",
xlab = "Size of keyword genesets",
ylab = "Number of keywords"
)
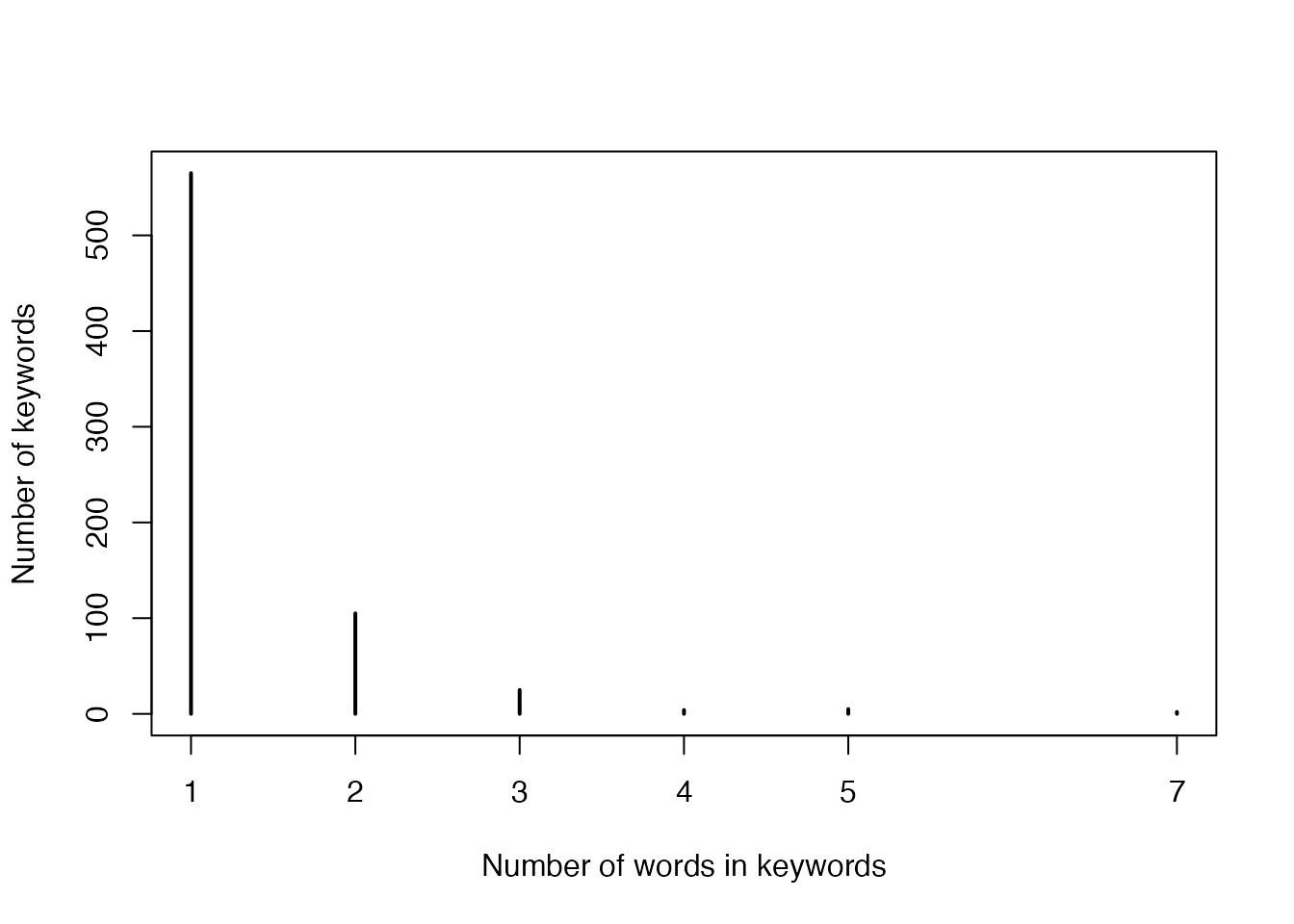
- Numbers of words in keywords:
plot(table(sapply(gregexpr(" |-|/", names(gl)), length)),
xlab = "Number of words in keywords",
ylab = "Number of keywords"
)
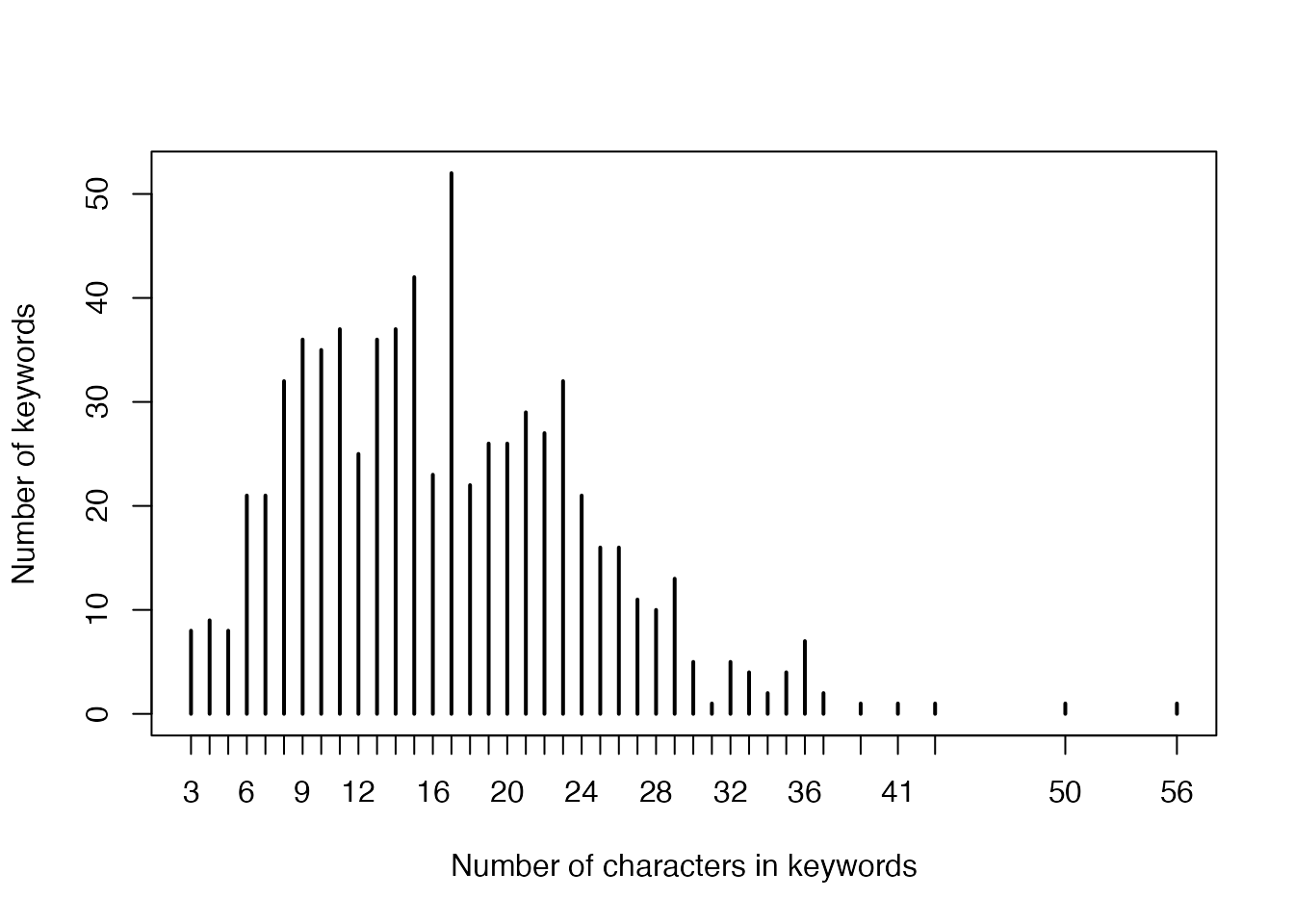
- Numbers of characters in keywords:
plot(table(nchar(names(gl))),
xlab = "Number of characters in keywords",
ylab = "Number of keywords"
)