Topic 2-03: Use clusterProfiler for ORA
Zuguang Gu z.gu@dkfz.de
2025-03-02
Source:vignettes/topic2_03_clusterProfiler_ora.Rmd
topic2_03_clusterProfiler_ora.RmdLet’s start with a vector of DE genes:
lt = readRDS(system.file("extdata", "ora.rds", package = "GSEAtraining"))
diff_gene = lt$diff_gene
bg_gene = lt$bg_gene
head(diff_gene)## [1] "FGR" "NIPAL3" "LAP3" "CASP10" "CAMKK1" "PRSS22"Load the clusterProfiler package.
clusterProfiler supports several major gene ID types in the input, but it is suggested to use Entrez IDs as the input because it is the “central gene ID type” in many databases/datasets.
We convert diff_gene to Entrez IDs. Note some genes may be lost due to the conversion.
library(GSEAtraining)
diff_gene = convert_to_entrez_id(diff_gene)## ## gene id might be SYMBOL (p = 0.980 )## 'select()' returned 1:many mapping between keys and columnshead(diff_gene)## FGR NIPAL3 LAP3 CASP10 CAMKK1 PRSS22
## "2268" "57185" "51056" "843" "84254" "64063"length(diff_gene)## [1] 835Next we perform ORA on different gene set sources.
GO enrichment
## ID Description GeneRatio
## GO:0002274 GO:0002274 myeloid leukocyte activation 51/779
## GO:0002237 GO:0002237 response to molecule of bacterial origin 63/779
## GO:0032496 GO:0032496 response to lipopolysaccharide 61/779
## GO:0050729 GO:0050729 positive regulation of inflammatory response 40/779
## GO:0050727 GO:0050727 regulation of inflammatory response 66/779
## GO:0002697 GO:0002697 regulation of immune effector process 63/779
## BgRatio RichFactor FoldEnrichment zScore pvalue
## GO:0002274 247/18986 0.2064777 5.032331 13.19430 5.149704e-22
## GO:0002237 375/18986 0.1680000 4.094542 12.51936 6.945328e-22
## GO:0032496 354/18986 0.1723164 4.199742 12.57018 8.355530e-22
## GO:0050729 162/18986 0.2469136 6.017845 13.26703 1.676133e-20
## GO:0050727 438/18986 0.1506849 3.672534 11.70487 3.037134e-20
## GO:0002697 410/18986 0.1536585 3.745008 11.62289 8.217123e-20
## p.adjust qvalue
## GO:0002274 1.409021e-18 1.099119e-18
## GO:0002237 1.409021e-18 1.099119e-18
## GO:0032496 1.409021e-18 1.099119e-18
## GO:0050729 2.119890e-17 1.653638e-17
## GO:0050727 3.072972e-17 2.397098e-17
## GO:0002697 6.928404e-17 5.404561e-17
## geneID
## GO:0002274 2268/7305/6556/22904/8692/3430/54209/50487/3696/3458/7474/9173/8807/8013/729230/3598/55509/6372/30817/5819/8291/3569/7097/2207/6283/80149/81501/6850/338339/11314/116071/6338/3965/5724/3576/136/712/8877/2624/2205/3579/2242/2204/5320/5359/728/2214/57126/10537/11027/6348
## GO:0002237 4843/6401/6556/50943/22904/5743/2920/3595/54209/3055/54/730249/5054/1440/6347/1594/57379/6648/942/7474/3552/29126/6590/6372/3553/10288/3620/948/3569/7097/10068/834/4283/6279/64332/133/4314/115362/10417/249/2634/9076/2921/6374/5196/2919/80149/717/64127/3965/3627/6373/5724/3576/2353/929/1051/2204/9516/1604/728/6891/11027
## GO:0032496 4843/6401/6556/50943/22904/5743/2920/3595/54209/3055/54/730249/5054/1440/6347/1594/57379/6648/942/7474/3552/29126/6590/6372/3553/10288/3620/948/3569/7097/10068/834/4283/6279/64332/133/4314/115362/10417/249/2634/9076/2921/6374/5196/2919/80149/717/64127/3965/3627/6373/5724/3576/2353/929/1051/2204/9516/1604/11027
## GO:0050729 8692/3430/5743/58484/54209/5008/50487/4210/5054/59341/3458/7474/9173/2633/729230/81030/3553/2867/3620/3569/7097/834/838/6279/64332/7941/2209/115362/2634/6280/6283/136/1051/4023/353514/5320/145741/3133/388125/6348
## GO:0050727 2268/6401/3082/50943/22904/8692/3430/55737/5743/58484/54209/5008/50487/25807/3055/54/730249/4210/5054/59341/3458/7474/8087/9173/2633/729230/7130/81030/3553/2867/3620/4907/3569/8870/7097/834/838/64116/90527/405753/6279/64332/7941/4314/2209/115362/56833/2634/6280/6283/6850/64127/136/2358/1051/6288/4023/8877/9021/338557/353514/5320/145741/3133/388125/6348
## GO:0002697 2268/4843/7305/50943/7037/2219/3383/54209/50487/54/3458/942/7474/57142/8809/8807/8013/29126/729230/3598/722/725/6372/3553/2867/30817/5819/948/3569/8547/64332/2209/26060/910/56833/10417/8741/6280/80149/6850/64127/11314/3965/5724/136/51237/3726/115727/64581/2624/2242/7293/441168/9516/1604/246778/6891/3107/3133/3134/57126/3106/387837
## Count
## GO:0002274 51
## GO:0002237 63
## GO:0032496 61
## GO:0050729 40
## GO:0050727 66
## GO:0002697 63KEGG enrichment
tb = enrichKEGG(gene = diff_gene, organism = "hsa")
head(tb)## category
## hsa04060 Environmental Information Processing
## hsa04061 Environmental Information Processing
## hsa04657 Organismal Systems
## hsa04380 Organismal Systems
## hsa04668 Environmental Information Processing
## hsa05323 Human Diseases
## subcategory ID
## hsa04060 Signaling molecules and interaction hsa04060
## hsa04061 Signaling molecules and interaction hsa04061
## hsa04657 Immune system hsa04657
## hsa04380 Development and regeneration hsa04380
## hsa04668 Signal transduction hsa04668
## hsa05323 Immune disease hsa05323
## Description
## hsa04060 Cytokine-cytokine receptor interaction
## hsa04061 Viral protein interaction with cytokine and cytokine receptor
## hsa04657 IL-17 signaling pathway
## hsa04380 Osteoclast differentiation
## hsa04668 TNF signaling pathway
## hsa05323 Rheumatoid arthritis
## GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
## hsa04060 50/452 298/8535 0.1677852 3.168246 9.009305 1.539612e-13
## hsa04061 27/452 100/8535 0.2700000 5.098341 9.748207 8.613618e-13
## hsa04657 25/452 95/8535 0.2631579 4.969143 9.199126 1.148639e-11
## hsa04380 29/452 143/8535 0.2027972 3.829368 8.068334 2.535667e-10
## hsa04668 26/452 119/8535 0.2184874 4.125641 8.119308 3.971319e-10
## hsa05323 23/452 95/8535 0.2421053 4.571612 8.277783 4.807896e-10
## p.adjust qvalue
## hsa04060 4.926759e-11 4.294708e-11
## hsa04061 1.378179e-10 1.201373e-10
## hsa04657 1.225215e-09 1.068033e-09
## hsa04380 2.028533e-08 1.768294e-08
## hsa04668 2.541644e-08 2.215578e-08
## hsa05323 2.564211e-08 2.235250e-08
## geneID
## hsa04060 53832/608/2920/3595/3589/5008/1440/6354/6347/55801/3458/3552/7850/9173/8809/8807/1441/729230/3624/3598/6372/3553/51554/9518/3601/3569/3557/4283/94/56477/58191/8741/3577/2921/6374/5196/2919/3627/6373/3576/8794/1237/3579/7293/8784/246778/6351/9560/6349/6348
## hsa04061 53832/2920/6354/6347/8809/8807/729230/6372/51554/3569/4283/56477/3577/2921/6374/5196/2919/3627/6373/3576/8794/1237/3579/6351/9560/6349/6348
## hsa04657 5743/2920/1440/6354/6347/3458/727897/6372/3553/2354/3569/4322/6279/3934/4314/6280/2921/6374/2919/3627/3576/2353/1051/8061/4312
## hsa04380 7305/54209/10326/54/11024/3458/3552/6772/2274/4688/8503/3553/2354/10288/7006/2212/2209/2215/6850/2353/126014/3726/8061/9021/353514/2214/11025/11027/79168
## hsa04668 843/64285/6401/5743/2920/3383/6347/8809/8503/6372/3659/3553/3569/4314/4323/8986/2921/6374/2919/64127/197259/3627/2353/3726/1051/9021
## hsa05323 2920/3383/3589/54/6347/10312/3458/942/3552/6372/3553/3569/7097/4314/8741/2921/6374/2919/3576/2353/4312/6349/6348
## Count
## hsa04060 50
## hsa04061 27
## hsa04657 25
## hsa04380 29
## hsa04668 26
## hsa05323 23Reactome enrichment
## ReactomePA v1.50.0 Learn more at https://yulab-smu.top/contribution-knowledge-mining/
##
## Please cite:
##
## Guangchuang Yu, Qing-Yu He. ReactomePA: an R/Bioconductor package for
## reactome pathway analysis and visualization. Molecular BioSystems.
## 2016, 12(2):477-479tb = enrichPathway(gene = diff_gene, organism = "human")
head(tb)## ID
## R-HSA-6798695 R-HSA-6798695
## R-HSA-6783783 R-HSA-6783783
## R-HSA-380108 R-HSA-380108
## R-HSA-449147 R-HSA-449147
## R-HSA-198933 R-HSA-198933
## R-HSA-6785807 R-HSA-6785807
## Description
## R-HSA-6798695 Neutrophil degranulation
## R-HSA-6783783 Interleukin-10 signaling
## R-HSA-380108 Chemokine receptors bind chemokines
## R-HSA-449147 Signaling by Interleukins
## R-HSA-198933 Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell
## R-HSA-6785807 Interleukin-4 and Interleukin-13 signaling
## GeneRatio BgRatio RichFactor FoldEnrichment zScore
## R-HSA-6798695 71/599 479/11146 0.1482255 2.758132 9.373182
## R-HSA-6783783 21/599 47/11146 0.4468085 8.314070 11.974436
## R-HSA-380108 22/599 59/11146 0.3728814 6.938457 10.898872
## R-HSA-449147 64/599 473/11146 0.1353066 2.517741 8.038480
## R-HSA-198933 27/599 133/11146 0.2030075 3.777499 7.679198
## R-HSA-6785807 23/599 108/11146 0.2129630 3.962747 7.373101
## pvalue p.adjust qvalue
## R-HSA-6798695 2.519471e-15 2.935183e-12 2.797938e-12
## R-HSA-6783783 5.190031e-15 3.023193e-12 2.881833e-12
## R-HSA-380108 1.099867e-13 4.271149e-11 4.071437e-11
## R-HSA-449147 4.612427e-12 1.343369e-09 1.280555e-09
## R-HSA-198933 1.607939e-09 3.746498e-07 3.571317e-07
## R-HSA-6785807 1.007665e-08 1.956549e-06 1.865064e-06
## geneID
## R-HSA-6798695 2268/64386/5329/7305/55/6556/2219/4680/4069/5836/10326/10562/3614/10493/10312/5768/5328/10549/7130/6590/27180/10288/1116/6947/948/10970/7097/6386/53831/2212/6279/3934/5795/2207/3101/2215/5284/1520/6282/6280/6283/3577/4332/2919/1508/338339/11314/6036/5724/929/126014/1084/2358/2357/116844/160364/10855/3310/4973/196527/3579/222487/199675/2204/1604/728/3107/57126/3106/150372/81567
## R-HSA-6783783 5743/2920/3383/7076/1440/6347/942/3552/7850/729230/3553/3569/3557/2919/3627/5724/3576/2357/6351/6349/6348
## R-HSA-380108 2920/6354/6347/729230/6372/51554/4283/56477/58191/3577/2921/6374/5196/2919/3627/6373/3576/1237/3579/6351/6349/6348
## R-HSA-449147 4843/53832/3082/6196/5743/2920/3595/3383/3589/5008/3055/7076/1440/6347/595/55801/3458/6648/942/3552/6772/7850/9173/8809/8807/8503/1441/729230/3598/3553/759/3601/948/7006/3569/3557/10068/834/1848/84166/3934/4314/6283/2919/6850/64127/3965/3627/5724/3576/2353/2357/3726/6288/9021/4582/4312/246778/5696/5699/5698/6351/6349/6348
## R-HSA-198933 7305/57823/1308/29992/3383/54209/11024/54210/27180/5819/10288/1280/2209/910/10871/11314/27036/126014/8519/342510/353514/2214/3107/3133/3134/3106/11027
## R-HSA-6785807 4843/3082/5743/3383/5008/7076/6347/595/3552/6772/3598/3553/948/3569/3934/4314/3576/2353/3726/6288/9021/4582/4312
## Count
## R-HSA-6798695 71
## R-HSA-6783783 21
## R-HSA-380108 22
## R-HSA-449147 64
## R-HSA-198933 27
## R-HSA-6785807 23DO enrichment
## DOSE v4.0.0 Learn more at https://yulab-smu.top/contribution-knowledge-mining/
##
## Please cite:
##
## Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an
## R/Bioconductor package for Disease Ontology Semantic and Enrichment
## analysis. Bioinformatics. 2015, 31(4):608-609## ID Description GeneRatio BgRatio
## DOID:934 DOID:934 viral infectious disease 69/410 336/7865
## DOID:0050161 DOID:0050161 lower respiratory tract disease 82/410 482/7865
## DOID:850 DOID:850 lung disease 70/410 367/7865
## DOID:1176 DOID:1176 bronchial disease 55/410 277/7865
## DOID:2841 DOID:2841 asthma 54/410 273/7865
## DOID:418 DOID:418 systemic scleroderma 31/410 94/7865
## RichFactor FoldEnrichment zScore pvalue p.adjust
## DOID:934 0.2053571 3.939351 12.91345 3.995847e-24 3.492370e-21
## DOID:0050161 0.1701245 3.263485 12.02748 6.172746e-23 2.697490e-20
## DOID:850 0.1907357 3.658869 12.23340 1.708620e-22 4.977780e-20
## DOID:1176 0.1985560 3.808884 11.16092 1.415077e-18 3.091944e-16
## DOID:2841 0.1978022 3.794425 11.02010 3.596275e-18 6.286288e-16
## DOID:418 0.3297872 6.326284 12.18259 1.810804e-17 2.637738e-15
## qvalue
## DOID:934 2.006336e-21
## DOID:0050161 1.549684e-20
## DOID:850 2.859690e-20
## DOID:1176 1.776295e-16
## DOID:2841 3.611417e-16
## DOID:418 1.515357e-15
## geneID
## DOID:934 6401/5329/3082/50943/5743/2920/5243/3383/51311/5054/768/1440/6354/6347/1594/3458/57379/6648/6772/9173/29126/729230/5328/722/725/6590/54210/6372/3659/3553/3620/3569/7097/4283/10410/2212/6279/4585/3934/58191/3577/2921/6374/2919/80149/54578/3627/3576/929/1051/713/627/9332/9021/4582/8784/5320/4312/246778/728/2214/6891/3107/8505/3106/6351/9560/6349/6348
## DOID:0050161 4843/5329/637/3082/50943/5743/2920/5243/3383/7076/4210/5054/259/1440/6354/6347/7450/4891/3458/6648/942/3552/6772/2744/9173/8809/727897/1490/1958/729230/5328/3598/6590/54210/3659/3553/1131/1116/9365/948/3569/3557/7097/10068/4322/834/4283/3067/2212/7941/3486/4314/10630/2921/6374/2919/64127/6890/6338/3627/6373/5724/3576/929/627/9332/2205/1237/3579/729238/4582/5320/4312/246778/728/2214/6891/3107/3133/3106/5698/6351
## DOID:850 4843/5329/637/3082/50943/5743/2920/3383/7076/4210/5054/259/1440/6354/6347/7450/4891/3458/6648/942/3552/6772/2744/9173/8809/727897/1490/1958/729230/5328/3598/6590/54210/3659/3553/1131/1116/9365/948/3569/3557/7097/10068/4322/4283/3486/4314/10630/2921/6374/2919/64127/6890/3627/6373/3576/929/627/9332/1237/3579/729238/4582/5320/4312/246778/728/2214/5698/6351
## DOID:1176 4843/50943/5743/5243/3383/4210/5054/259/1440/6354/6347/3458/6648/942/3552/6772/9173/8809/727897/1958/729230/5328/3598/3659/3553/1131/1116/3569/3557/7097/834/3067/2212/7941/6374/2919/64127/6890/6338/3627/3576/929/627/2205/1237/729238/4582/246778/728/2214/6891/3107/3133/3106/6351
## DOID:2841 4843/50943/5743/5243/3383/4210/5054/259/1440/6354/6347/3458/6648/942/3552/6772/9173/8809/727897/1958/729230/5328/3598/3659/3553/1131/1116/3569/3557/7097/834/3067/2212/7941/6374/2919/64127/6890/3627/3576/929/627/2205/1237/729238/4582/246778/728/2214/6891/3107/3133/3106/6351
## DOID:418 6556/3082/5743/3383/5054/6347/7450/3458/942/3553/1116/3569/7097/4283/2212/3486/4314/58191/6374/5196/308/6890/3627/627/4582/4312/246778/6891/629/6351/6348
## Count
## DOID:934 69
## DOID:0050161 82
## DOID:850 70
## DOID:1176 55
## DOID:2841 54
## DOID:418 31MSigDB enrichment
There is no built-in function specific for MSigDB gene sets, but there is a universal function enricher() which accepts manually-specified gene sets. The gene sets object is simply a two-column data frame:
- the first column is the gene set ID
- the second column is the gene ID
library(msigdbr)
gene_sets = msigdbr(category = "H")
map = gene_sets[, c("gs_name", "entrez_gene")]
head(map)## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 HALLMARK_ADIPOGENESIS 19
## 2 HALLMARK_ADIPOGENESIS 11194
## 3 HALLMARK_ADIPOGENESIS 10449
## 4 HALLMARK_ADIPOGENESIS 33
## 5 HALLMARK_ADIPOGENESIS 34
## 6 HALLMARK_ADIPOGENESIS 35## ID
## HALLMARK_INTERFERON_GAMMA_RESPONSE HALLMARK_INTERFERON_GAMMA_RESPONSE
## HALLMARK_INFLAMMATORY_RESPONSE HALLMARK_INFLAMMATORY_RESPONSE
## HALLMARK_INTERFERON_ALPHA_RESPONSE HALLMARK_INTERFERON_ALPHA_RESPONSE
## HALLMARK_TNFA_SIGNALING_VIA_NFKB HALLMARK_TNFA_SIGNALING_VIA_NFKB
## HALLMARK_COMPLEMENT HALLMARK_COMPLEMENT
## HALLMARK_IL6_JAK_STAT3_SIGNALING HALLMARK_IL6_JAK_STAT3_SIGNALING
## Description GeneRatio
## HALLMARK_INTERFERON_GAMMA_RESPONSE HALLMARK_INTERFERON_GAMMA_RESPONSE 55/347
## HALLMARK_INFLAMMATORY_RESPONSE HALLMARK_INFLAMMATORY_RESPONSE 54/347
## HALLMARK_INTERFERON_ALPHA_RESPONSE HALLMARK_INTERFERON_ALPHA_RESPONSE 33/347
## HALLMARK_TNFA_SIGNALING_VIA_NFKB HALLMARK_TNFA_SIGNALING_VIA_NFKB 49/347
## HALLMARK_COMPLEMENT HALLMARK_COMPLEMENT 39/347
## HALLMARK_IL6_JAK_STAT3_SIGNALING HALLMARK_IL6_JAK_STAT3_SIGNALING 22/347
## BgRatio RichFactor FoldEnrichment zScore
## HALLMARK_INTERFERON_GAMMA_RESPONSE 200/4383 0.2750000 3.473559 10.498293
## HALLMARK_INFLAMMATORY_RESPONSE 200/4383 0.2700000 3.410403 10.230248
## HALLMARK_INTERFERON_ALPHA_RESPONSE 97/4383 0.3402062 4.297186 9.627835
## HALLMARK_TNFA_SIGNALING_VIA_NFKB 200/4383 0.2450000 3.094625 8.890021
## HALLMARK_COMPLEMENT 200/4383 0.1950000 2.463069 6.209566
## HALLMARK_IL6_JAK_STAT3_SIGNALING 87/4383 0.2528736 3.194077 6.060448
## pvalue p.adjust qvalue
## HALLMARK_INTERFERON_GAMMA_RESPONSE 1.283631e-17 6.033064e-16 4.594047e-16
## HALLMARK_INFLAMMATORY_RESPONSE 6.465947e-17 1.519498e-15 1.157064e-15
## HALLMARK_INTERFERON_ALPHA_RESPONSE 8.600159e-14 1.347358e-12 1.025984e-12
## HALLMARK_TNFA_SIGNALING_VIA_NFKB 1.359501e-13 1.597414e-12 1.216396e-12
## HALLMARK_COMPLEMENT 6.053975e-08 5.690736e-07 4.333372e-07
## HALLMARK_IL6_JAK_STAT3_SIGNALING 5.774922e-07 4.523689e-06 3.444690e-06
## geneID
## HALLMARK_INTERFERON_GAMMA_RESPONSE 51056/57823/10797/3430/5743/3383/10135/6354/6347/6648/942/6772/29126/7130/81030/3659/3620/6737/94240/3601/8638/10906/3569/10068/834/10561/4283/7453/5371/84166/10410/2209/81894/9246/115361/3429/116071/6890/10791/3627/6373/2357/3669/54625/219285/9021/10581/7127/5359/5696/5699/10437/3106/5698/629
## HALLMARK_INFLAMMATORY_RESPONSE 6401/5329/2769/490/3383/5008/7076/4210/366/3249/6324/10135/3696/5054/1440/6354/6347/4891/1839/3552/8809/8807/1441/3624/7130/3759/6372/3659/3553/2867/3601/3569/7097/9153/4283/7162/133/4323/10630/60675/64127/3627/6373/5724/3576/136/929/2357/4973/8877/8519/1604/3269/728
## HALLMARK_INTERFERON_ALPHA_RESPONSE 51056/3430/135112/3659/6737/94240/8638/10906/2766/834/10561/83666/7453/10410/81894/9246/2634/115361/3429/116071/6890/3627/6373/3669/54625/219285/10581/8519/5359/5696/3107/10437/5698
## HALLMARK_TNFA_SIGNALING_VIA_NFKB 5329/490/5743/2920/3383/10135/5054/6347/374/595/6648/1839/3552/5341/8013/1958/3624/5328/7130/50486/6372/3659/3553/2354/3601/3569/8870/7097/3491/4929/467/2921/2919/80149/6890/3627/6373/2353/3726/1051/4973/8061/8877/7262/9021/23764/7127/9516/6351
## HALLMARK_COMPLEMENT 51056/843/5329/2219/5251/7076/760/7980/5054/5341/725/3659/948/3569/4322/834/838/1848/7941/4323/2207/714/1520/6280/6283/2919/308/1508/717/255738/1051/712/4973/23764/5359/1604/5698/629/4321
## HALLMARK_IL6_JAK_STAT3_SIGNALING 6354/5967/6772/7850/8809/1441/3659/3553/3601/948/3569/7097/4283/94/2921/5196/2919/3627/6373/929/9021/5320
## Count
## HALLMARK_INTERFERON_GAMMA_RESPONSE 55
## HALLMARK_INFLAMMATORY_RESPONSE 54
## HALLMARK_INTERFERON_ALPHA_RESPONSE 33
## HALLMARK_TNFA_SIGNALING_VIA_NFKB 49
## HALLMARK_COMPLEMENT 39
## HALLMARK_IL6_JAK_STAT3_SIGNALING 22Setting background
Note, in clusterProfiler, the background is intersect(all genes in the gene sets, user’s background genes).
bg_gene = convert_to_entrez_id(bg_gene)## gene id might be SYMBOL (p = 0.980 )## 'select()' returned 1:many mapping between keys and columnsbg_gene = sample(bg_gene, 10000)
tb = enrichGO(gene = diff_gene, ont = "BP", OrgDb = org.Hs.eg.db, universe = bg_gene)
head(tb)## ID Description
## GO:0002274 GO:0002274 myeloid leukocyte activation
## GO:0006954 GO:0006954 inflammatory response
## GO:0032496 GO:0032496 response to lipopolysaccharide
## GO:0002237 GO:0002237 response to molecule of bacterial origin
## GO:0009617 GO:0009617 response to bacterium
## GO:0002275 GO:0002275 myeloid cell activation involved in immune response
## GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
## GO:0002274 31/779 123/8993 0.2520325 2.909536 6.566563 3.400639e-08
## GO:0006954 68/779 426/8993 0.1596244 1.842750 5.487933 3.945138e-07
## GO:0032496 34/779 162/8993 0.2098765 2.422875 5.627803 8.611459e-07
## GO:0002237 34/779 169/8993 0.2011834 2.322519 5.344809 2.375508e-06
## GO:0009617 56/779 347/8993 0.1613833 1.863055 5.049114 3.084574e-06
## GO:0002275 16/779 51/8993 0.3137255 3.621737 5.781978 3.421354e-06
## p.adjust qvalue
## GO:0002274 0.0001418067 0.0001418067
## GO:0006954 0.0008225613 0.0008225613
## GO:0032496 0.0011969928 0.0011969928
## GO:0002237 0.0020381496 0.0020381496
## GO:0009617 0.0020381496 0.0020381496
## GO:0002275 0.0020381496 0.0020381496
## geneID
## GO:0002274 2268/7305/22904/8692/3430/50487/3696/7474/8807/8013/3598/30817/5819/8291/6283/80149/6850/338339/11314/116071/3965/5724/3576/8877/2624/2205/3579/5320/5359/2214/11027
## GO:0006954 2268/4843/6401/7305/22904/8692/3430/55737/5743/4069/58484/50487/25807/3055/730249/4210/6347/10312/7474/8087/3552/57142/8807/81030/2867/30817/6737/6289/8870/834/2212/6279/133/4314/115362/26060/8986/2634/6283/60675/2921/6374/80149/6850/3965/6373/5724/3576/2353/929/2358/2357/1051/4973/4023/8877/2205/3579/9021/7293/353514/5320/5359/246778/2214/388125/150372/6349
## GO:0032496 4843/6401/22904/5743/3055/730249/6347/1594/57379/7474/3552/29126/6590/10288/10068/834/6279/133/4314/115362/10417/2634/2921/6374/80149/717/3965/6373/5724/3576/2353/929/1051/11027
## GO:0002237 4843/6401/22904/5743/3055/730249/6347/1594/57379/7474/3552/29126/6590/10288/10068/834/6279/133/4314/115362/10417/2634/2921/6374/80149/717/3965/6373/5724/3576/2353/929/1051/11027
## GO:0009617 2268/4843/6401/22904/5743/4069/58484/3055/730249/6347/54757/1594/57379/7474/3552/29126/722/5266/6590/10288/3512/10068/4316/834/10561/6279/3934/133/4314/115362/10417/2634/115361/6283/2921/6374/80149/6850/717/338339/3965/6373/5724/3576/2353/929/2358/116844/115727/1051/4023/5320/246778/3106/11027/358
## GO:0002275 2268/7305/22904/3430/50487/8013/3598/30817/8291/6850/11314/3965/5724/2624/2205/11027
## Count
## GO:0002274 31
## GO:0006954 68
## GO:0032496 34
## GO:0002237 34
## GO:0009617 56
## GO:0002275 16Warning: It seems when setting
universe, the input gene listdiff_geneis not intersected touniversein the analysis. You can check the values inGeneRatiowhich is 777 for all DE genes, and this number is the same as whenuniverseis not manually set. So be careful with this “improper” behaviour.
We can compare ORA with different backgrounds:
tb1 = enrichGO(gene = diff_gene, ont = "BP", OrgDb = org.Hs.eg.db)
tb2 = enrichGO(gene = diff_gene, ont = "BP", OrgDb = org.Hs.eg.db, universe = bg_gene)
tb1 = tb1@result
tb2 = tb2@result
cn = intersect(tb1$ID, tb2$ID)
plot(tb1[cn, "pvalue"], tb2[cn, "pvalue"], log = "xy",
xlim = c(1e-25, 1), ylim = c(1e-25, 1),
xlab = "default background", ylab = "self-defined background")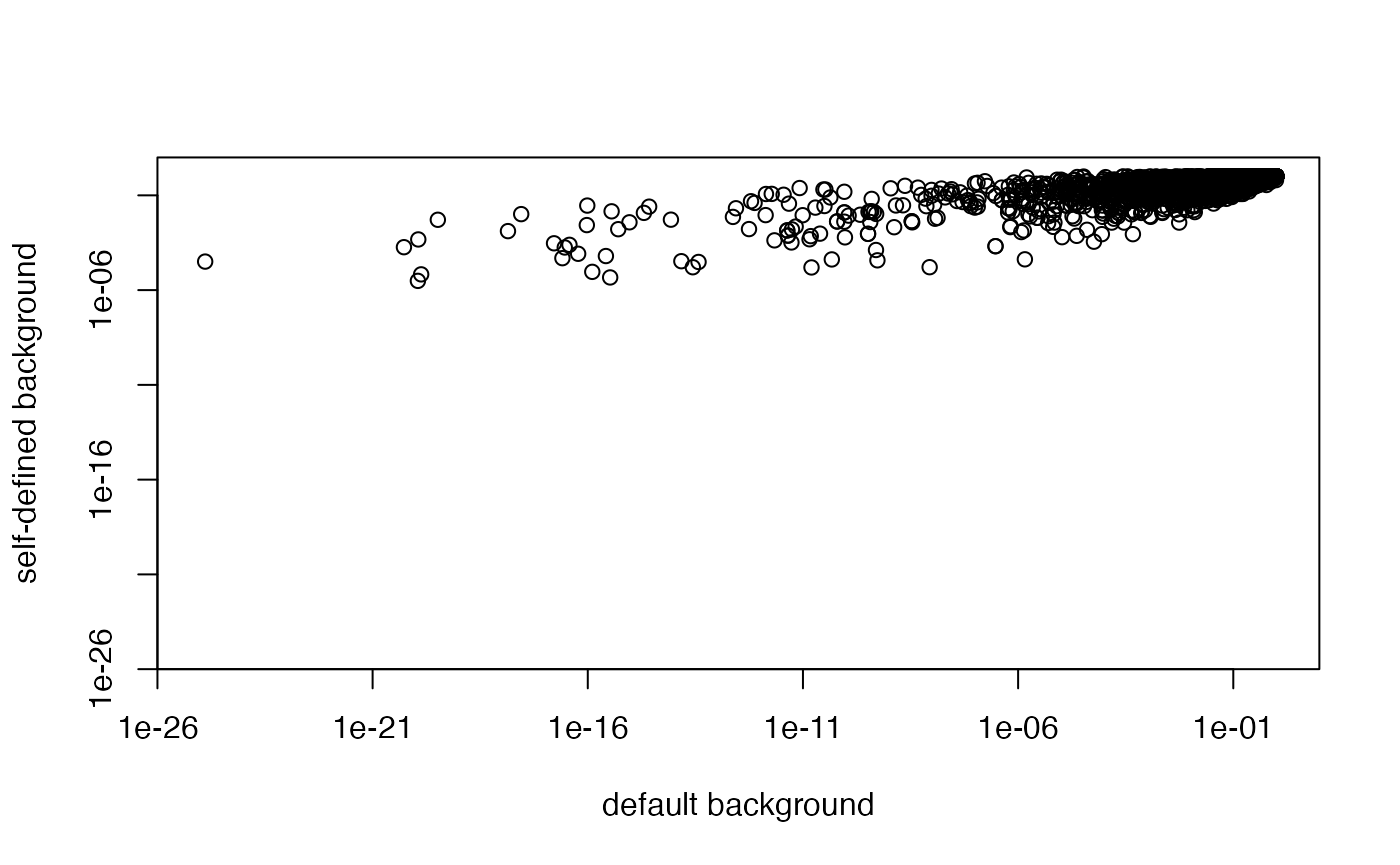
Or overlap between the significant GO terms:
library(eulerr)
plot(euler(list("default_background" = tb1$ID[tb1$p.adjust < 0.05],
"bg_gene" = tb2$ID[tb2$p.adjust < 0.05])),
quantities = TRUE)
Example for other organism
- GO enrichment
enrichGO() accepts an OrgDb object as the source of the GO gene sets. We take pig as an example. random_genes() is from GSEAtraining package.
library(org.Ss.eg.db)## diff_gene = random_genes(org.Ss.eg.db, 1000, "ENTREZID")## 'select()' returned 1:many mapping between keys and columnsHere we set pvalueCutoff = 1, qvalueCutoff = 1 because genes are random genes and it is expected there won’t be too much significant terms left.
tb = enrichGO(gene = diff_gene, ont = "BP", OrgDb = org.Ss.eg.db,
pvalueCutoff = 1, qvalueCutoff = 1)
head(tb)## ID
## GO:0046686 GO:0046686
## GO:0071276 GO:0071276
## GO:1901137 GO:1901137
## GO:0009100 GO:0009100
## GO:0042058 GO:0042058
## GO:0043368 GO:0043368
## Description
## GO:0046686 response to cadmium ion
## GO:0071276 cellular response to cadmium ion
## GO:1901137 carbohydrate derivative biosynthetic process
## GO:0009100 glycoprotein metabolic process
## GO:0042058 regulation of epidermal growth factor receptor signaling pathway
## GO:0043368 positive T cell selection
## GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
## GO:0046686 4/394 10/7920 0.4000000 8.040609 5.097100 0.0009969006
## GO:0071276 4/394 10/7920 0.4000000 8.040609 5.097100 0.0009969006
## GO:1901137 23/394 244/7920 0.0942623 1.894816 3.248345 0.0023067425
## GO:0009100 16/394 152/7920 0.1052632 2.115950 3.178433 0.0035629169
## GO:0042058 4/394 14/7920 0.2857143 5.743292 4.064121 0.0040541352
## GO:0043368 4/394 15/7920 0.2666667 5.360406 3.867433 0.0053141264
## p.adjust qvalue
## GO:0046686 0.9876963 0.9876963
## GO:0071276 0.9876963 0.9876963
## GO:1901137 0.9876963 0.9876963
## GO:0009100 0.9876963 0.9876963
## GO:0042058 0.9876963 0.9876963
## GO:0043368 0.9876963 0.9876963
## geneID
## GO:0046686 396609/397417/396610/100127139
## GO:0071276 396609/397417/396610/100127139
## GO:1901137 397385/414851/100627727/100157880/100512485/100517301/397599/100124379/100523821/100620659/100156098/100520822/100525767/100157902/100524201/100522057/100738545/100512094/100520587/100233182/397634/100522426/100155947
## GO:0009100 397385/414851/100627727/397599/100124379/396926/100620659/100156098/100525767/733672/100524201/100522057/100738545/397634/100522426/100155947
## GO:0042058 100624229/100154116/100157266/100518854
## GO:0043368 100155546/396926/100155958/733648
## Count
## GO:0046686 4
## GO:0071276 4
## GO:1901137 23
## GO:0009100 16
## GO:0042058 4
## GO:0043368 4- KEGG enrichment
The KEGG code of a specific organism can be found at https://rest.kegg.jp/list/organism
tb = enrichKEGG(gene = diff_gene, organism = "ssc", pvalueCutoff = 1, qvalueCutoff = 1)
head(tb)## category subcategory ID
## ssc00040 Metabolism Carbohydrate metabolism ssc00040
## ssc04662 Organismal Systems Immune system ssc04662
## ssc05167 Human Diseases Infectious disease: viral ssc05167
## ssc04210 Cellular Processes Cell growth and death ssc04210
## ssc04215 Cellular Processes Cell growth and death ssc04215
## ssc00330 Metabolism Amino acid metabolism ssc00330
## Description GeneRatio BgRatio
## ssc00040 Pentose and glucuronate interconversions 5/415 26/9198
## ssc04662 B cell receptor signaling pathway 9/415 84/9198
## ssc05167 Kaposi sarcoma-associated herpesvirus infection 16/415 196/9198
## ssc04210 Apoptosis 12/415 135/9198
## ssc04215 Apoptosis - multiple species 5/415 35/9198
## ssc00330 Arginine and proline metabolism 6/415 50/9198
## RichFactor FoldEnrichment zScore pvalue p.adjust qvalue
## ssc00040 0.19230769 4.262280 3.620775 0.005472404 0.8368088 0.8320657
## ssc04662 0.10714286 2.374699 2.751176 0.013094621 0.8368088 0.8320657
## ssc05167 0.08163265 1.809294 2.489377 0.015705317 0.8368088 0.8320657
## ssc04210 0.08888889 1.970120 2.468211 0.018745757 0.8368088 0.8320657
## ssc04215 0.14285714 3.166265 2.790953 0.019394478 0.8368088 0.8320657
## ssc00330 0.12000000 2.659663 2.557805 0.024134925 0.8368088 0.8320657
## geneID
## ssc00040 100627727/100525819/397337/100623255/396816
## ssc04662 100521529/100518663/100511898/100125346/780427/100737448/100134969/100522792/100518251
## ssc05167 100518663/100135029/100151828/100512852/100519790/396826/396609/100170126/100511902/100737448/100134969/100526085/100518251/733648/396610/100155666
## ssc04210 100515575/100510978/396926/396826/396609/100737448/397266/100622769/100049693/100518917/100518251/396610
## ssc04215 100515575/396609/397266/110257143/396610
## ssc00330 397264/100037299/100512885/100153961/397557/396968
## Count
## ssc00040 5
## ssc04662 9
## ssc05167 16
## ssc04210 12
## ssc04215 5
## ssc00330 6- MSigDB
Use msigdbr::msigdbr_species() to see which organisms are supported.
gene_sets = msigdbr(species = "pig", category = "H")
map = gene_sets[, c("gs_name", "entrez_gene")]
map$entrez_gene = as.character(map$entrez_gene)
tb = enricher(gene = diff_gene, TERM2GENE = map, pvalueCutoff = 1, qvalueCutoff = 1)
head(tb)## ID
## HALLMARK_MYOGENESIS HALLMARK_MYOGENESIS
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION
## HALLMARK_PI3K_AKT_MTOR_SIGNALING HALLMARK_PI3K_AKT_MTOR_SIGNALING
## HALLMARK_CHOLESTEROL_HOMEOSTASIS HALLMARK_CHOLESTEROL_HOMEOSTASIS
## HALLMARK_GLYCOLYSIS HALLMARK_GLYCOLYSIS
## HALLMARK_MITOTIC_SPINDLE HALLMARK_MITOTIC_SPINDLE
## Description
## HALLMARK_MYOGENESIS HALLMARK_MYOGENESIS
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION
## HALLMARK_PI3K_AKT_MTOR_SIGNALING HALLMARK_PI3K_AKT_MTOR_SIGNALING
## HALLMARK_CHOLESTEROL_HOMEOSTASIS HALLMARK_CHOLESTEROL_HOMEOSTASIS
## HALLMARK_GLYCOLYSIS HALLMARK_GLYCOLYSIS
## HALLMARK_MITOTIC_SPINDLE HALLMARK_MITOTIC_SPINDLE
## GeneRatio BgRatio RichFactor
## HALLMARK_MYOGENESIS 14/204 192/4191 0.07291667
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 14/204 194/4191 0.07216495
## HALLMARK_PI3K_AKT_MTOR_SIGNALING 8/204 103/4191 0.07766990
## HALLMARK_CHOLESTEROL_HOMEOSTASIS 6/204 72/4191 0.08333333
## HALLMARK_GLYCOLYSIS 13/204 196/4191 0.06632653
## HALLMARK_MITOTIC_SPINDLE 13/204 196/4191 0.06632653
## FoldEnrichment zScore pvalue
## HALLMARK_MYOGENESIS 1.498009 1.597757 0.08238532
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 1.482565 1.556642 0.08797199
## HALLMARK_PI3K_AKT_MTOR_SIGNALING 1.595660 1.384396 0.12688504
## HALLMARK_CHOLESTEROL_HOMEOSTASIS 1.712010 1.378337 0.13621701
## HALLMARK_GLYCOLYSIS 1.362620 1.176036 0.15653848
## HALLMARK_MITOTIC_SPINDLE 1.362620 1.176036 0.15653848
## p.adjust qvalue
## HALLMARK_MYOGENESIS 0.9940567 0.9940567
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 0.9940567 0.9940567
## HALLMARK_PI3K_AKT_MTOR_SIGNALING 0.9940567 0.9940567
## HALLMARK_CHOLESTEROL_HOMEOSTASIS 0.9940567 0.9940567
## HALLMARK_GLYCOLYSIS 0.9940567 0.9940567
## HALLMARK_MITOTIC_SPINDLE 0.9940567 0.9940567
## geneID
## HALLMARK_MYOGENESIS 397264/100049656/100736818/100302506/397667/100151828/100520244/100739757/100153623/100155338/100101928/100049693/100522432/100155578
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 100154530/100738910/100302506/100624264/397532/396826/100155607/100519746/100521017/100525583/396769/397092/100515628/100048956
## HALLMARK_PI3K_AKT_MTOR_SIGNALING 100624229/100519790/396609/100737448/100522792/100523972/100620726/396610
## HALLMARK_CHOLESTEROL_HOMEOSTASIS 100152230/100144419/100521017/397269/397561/100155380
## HALLMARK_GLYCOLYSIS 397136/100233197/100037299/100514793/733639/100525583/100514523/100512094/100621300/397628/396968/100156052/553107
## HALLMARK_MITOTIC_SPINDLE 100518983/100517117/100625051/100523340/100514649/100626498/397266/100512860/100516187/100049693/100738172/100624059/100153677
## Count
## HALLMARK_MYOGENESIS 14
## HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 14
## HALLMARK_PI3K_AKT_MTOR_SIGNALING 8
## HALLMARK_CHOLESTEROL_HOMEOSTASIS 6
## HALLMARK_GLYCOLYSIS 13
## HALLMARK_MITOTIC_SPINDLE 13Examples for less well-studied organisms
When an OrgDb object is avaiable
You can use enrichGO() as long as you have an OrgDb object:
Taking dolphin as an example.
library(AnnotationHub)## Loading required package: BiocFileCache## Loading required package: dbplyr##
## Attaching package: 'AnnotationHub'## The following object is masked from 'package:Biobase':
##
## cacheah = AnnotationHub()
org_db = ah[["AH118180"]]## loading from cachediff_gene = random_genes(org_db, 1000, "ENTREZID")
tb = enrichGO(diff_gene, ont = "BP", OrgDb = org_db,
pvalueCutoff = 1, qvalueCutoff = 1)
head(tb)## ID Description
## GO:0006188 GO:0006188 IMP biosynthetic process
## GO:0032543 GO:0032543 mitochondrial translation
## GO:0071900 GO:0071900 regulation of protein serine/threonine kinase activity
## GO:0120036 GO:0120036 plasma membrane bounded cell projection organization
## GO:0006473 GO:0006473 protein acetylation
## GO:0046040 GO:0046040 IMP metabolic process
## GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
## GO:0006188 3/453 10/13726 0.30000000 9.090066 4.727800 0.003603571
## GO:0032543 5/453 33/13726 0.15151515 4.590943 3.815372 0.004236004
## GO:0071900 7/453 66/13726 0.10606061 3.213660 3.330261 0.005916926
## GO:0120036 26/453 473/13726 0.05496829 1.665551 2.721295 0.007733292
## GO:0006473 3/453 13/13726 0.23076923 6.992359 3.993227 0.007978294
## GO:0046040 3/453 13/13726 0.23076923 6.992359 3.993227 0.007978294
## p.adjust qvalue
## GO:0006188 0.943299 0.943299
## GO:0032543 0.943299 0.943299
## GO:0071900 0.943299 0.943299
## GO:0120036 0.943299 0.943299
## GO:0006473 0.943299 0.943299
## GO:0046040 0.943299 0.943299
## geneID
## GO:0006188 101315901/101322662/101323582
## GO:0032543 101325303/101324614/101327888/101317961/101328773
## GO:0071900 101330246/101325657/117313024/117308306/101338481/101321085/101334943
## GO:0120036 101315722/101332080/117310101/101337789/101335043/101327264/101333027/101335978/101331724/101329010/101322494/101319665/101330777/101337069/101323465/101321040/101339730/101338906/101321994/101334024/101322276/101319062/101336302/109547263/101317632/117308159
## GO:0006473 101322089/101319574/117309525
## GO:0046040 101315901/101322662/101323582
## Count
## GO:0006188 3
## GO:0032543 5
## GO:0071900 7
## GO:0120036 26
## GO:0006473 3
## GO:0046040 3enrichKEGG() also supports many other orgainsms
enrichKEGG(gene = diff_gene, organism = ...)Manually construct the gene sets
We have introduced many ways to obtain gene sets for less well-studies organisms. The only thing to do here is to convert gene sets to a two-column data frame where gene sets are in the first column and genes are in the second column. Then use enricher().
enricher(diff_gene, TERM2GENE = ...)Look at the tb object
We used the same variable name tb for the object returned by the various enrichment functions. They are all in the same format. It looks like a table, but be careful, it is actually not:
class(tb)## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"str(tb)## Formal class 'enrichResult' [package "DOSE"] with 15 slots
## ..@ result :'data.frame': 1459 obs. of 12 variables:
## .. ..$ ID : chr [1:1459] "GO:0006188" "GO:0032543" "GO:0071900" "GO:0120036" ...
## .. ..$ Description : chr [1:1459] "IMP biosynthetic process" "mitochondrial translation" "regulation of protein serine/threonine kinase activity" "plasma membrane bounded cell projection organization" ...
## .. ..$ GeneRatio : chr [1:1459] "3/453" "5/453" "7/453" "26/453" ...
## .. ..$ BgRatio : chr [1:1459] "10/13726" "33/13726" "66/13726" "473/13726" ...
## .. ..$ RichFactor : num [1:1459] 0.3 0.152 0.106 0.055 0.231 ...
## .. ..$ FoldEnrichment: num [1:1459] 9.09 4.59 3.21 1.67 6.99 ...
## .. ..$ zScore : num [1:1459] 4.73 3.82 3.33 2.72 3.99 ...
## .. ..$ pvalue : num [1:1459] 0.0036 0.00424 0.00592 0.00773 0.00798 ...
## .. ..$ p.adjust : num [1:1459] 0.943 0.943 0.943 0.943 0.943 ...
## .. ..$ qvalue : num [1:1459] 0.943 0.943 0.943 0.943 0.943 ...
## .. ..$ geneID : chr [1:1459] "101315901/101322662/101323582" "101325303/101324614/101327888/101317961/101328773" "101330246/101325657/117313024/117308306/101338481/101321085/101334943" "101315722/101332080/117310101/101337789/101335043/101327264/101333027/101335978/101331724/101329010/101322494/1"| __truncated__ ...
## .. ..$ Count : int [1:1459] 3 5 7 26 3 3 3 26 11 3 ...
## ..@ pvalueCutoff : num 1
## ..@ pAdjustMethod: chr "BH"
## ..@ qvalueCutoff : num 1
## ..@ organism : chr "Tursiops truncatus"
## ..@ ontology : chr "BP"
## ..@ gene : chr [1:1000] "101324925" "117312870" "117308376" "101333535" ...
## ..@ keytype : chr "ENTREZID"
## ..@ universe : chr [1:13726] "101315666" "101325920" "101326047" "101328885" ...
## ..@ gene2Symbol : chr(0)
## ..@ geneSets :List of 1980
## .. ..$ GO:0000002: chr [1:10] "101315666" "101325920" "101326047" "101328885" ...
## .. ..$ GO:0000027: chr [1:33] "101315465" "101316856" "101319900" "101320222" ...
## .. ..$ GO:0000028: chr [1:34] "101317415" "101318043" "101318467" "101318567" ...
## .. ..$ GO:0000041: chr [1:87] "101315672" "101316332" "101316680" "101316885" ...
## .. ..$ GO:0000045: chr [1:44] "101316021" "101316129" "101317161" "101317562" ...
## .. ..$ GO:0000070: chr [1:69] "101315654" "101315716" "101316211" "101316537" ...
## .. ..$ GO:0000075: chr [1:63] "101316695" "101316733" "101318624" "101319304" ...
## .. ..$ GO:0000076: chr [1:13] "101316733" "101320717" "101320935" "101323238" ...
## .. ..$ GO:0000077: chr [1:31] "101316733" "101319304" "101320252" "101320465" ...
## .. ..$ GO:0000079: chr [1:38] "101316500" "101317117" "101319016" "101319831" ...
## .. ..$ GO:0000086: chr [1:34] "101316733" "101318219" "101318851" "101319304" ...
## .. ..$ GO:0000096: chr [1:19] "101317866" "101318072" "101318630" "101320181" ...
## .. ..$ GO:0000097: chr [1:14] "101318072" "101318630" "101320181" "101320414" ...
## .. ..$ GO:0000122: chr [1:289] "101315507" "101315608" "101315627" "101315712" ...
## .. ..$ GO:0000154: chr [1:30] "101315879" "101316217" "101318617" "101318650" ...
## .. ..$ GO:0000165: chr [1:150] "101315540" "101315821" "101315844" "101315877" ...
## .. ..$ GO:0000184: chr [1:17] "101317466" "101317816" "101318077" "101320986" ...
## .. ..$ GO:0000209: chr [1:107] "101315684" "101315840" "101316076" "101316166" ...
## .. ..$ GO:0000226: chr [1:319] "101315473" "101315515" "101315532" "101315537" ...
## .. ..$ GO:0000245: chr [1:28] "101316036" "101316848" "101318225" "101318825" ...
## .. ..$ GO:0000278: chr [1:306] "101315654" "101315716" "101315741" "101315815" ...
## .. ..$ GO:0000280: chr [1:154] "101315650" "101315654" "101315716" "101316211" ...
## .. ..$ GO:0000288: chr [1:24] "101315620" "101315678" "101317451" "101317568" ...
## .. ..$ GO:0000289: chr [1:14] "101315678" "101317451" "101317568" "101318606" ...
## .. ..$ GO:0000302: chr [1:16] "101319210" "101319583" "101322539" "101323139" ...
## .. ..$ GO:0000375: chr [1:230] "101315583" "101315667" "101315762" "101315834" ...
## .. ..$ GO:0000377: chr [1:230] "101315583" "101315667" "101315762" "101315834" ...
## .. ..$ GO:0000380: chr [1:45] "101315762" "101316036" "101317431" "101317847" ...
## .. ..$ GO:0000381: chr [1:34] "101315762" "101317847" "101319370" "101320027" ...
## .. ..$ GO:0000398: chr [1:230] "101315583" "101315667" "101315762" "101315834" ...
## .. ..$ GO:0000413: chr [1:29] "101316348" "101317534" "101318791" "101321235" ...
## .. ..$ GO:0000422: chr [1:33] "101316021" "101317608" "101317975" "101318574" ...
## .. ..$ GO:0000460: chr [1:32] "101318334" "101318462" "101318699" "101319173" ...
## .. ..$ GO:0000462: chr [1:33] "101316911" "101317350" "101317639" "101318699" ...
## .. ..$ GO:0000463: chr [1:17] "101315781" "101320270" "101320916" "101321745" ...
## .. ..$ GO:0000466: chr [1:23] "101318334" "101318699" "101319173" "101320141" ...
## .. ..$ GO:0000470: chr [1:24] "101315781" "101318617" "101320270" "101320916" ...
## .. ..$ GO:0000731: chr [1:16] "101316946" "101319167" "101321519" "101323763" ...
## .. ..$ GO:0000819: chr [1:73] "101315654" "101315716" "101316211" "101316537" ...
## .. ..$ GO:0000902: chr [1:253] "101315541" "101315722" "101316024" "101316049" ...
## .. ..$ GO:0000910: chr [1:64] "101315625" "101315654" "101315925" "101316479" ...
## .. ..$ GO:0000956: chr [1:59] "101315620" "101315678" "101315917" "101316856" ...
## .. ..$ GO:0000959: chr [1:16] "101315630" "101317316" "101317961" "101319361" ...
## .. ..$ GO:0000963: chr [1:12] "101317863" "101317961" "101319361" "101319586" ...
## .. ..$ GO:0000964: chr [1:2] "101317961" "101319586"
## .. ..$ GO:0000966: chr [1:23] "101317961" "101318647" "101318699" "101319586" ...
## .. ..$ GO:0001101: chr [1:17] "101316372" "101316666" "101319583" "101320291" ...
## .. ..$ GO:0001192: chr "101327079"
## .. ..$ GO:0001193: chr "101327079"
## .. ..$ GO:0001501: chr [1:33] "101315794" "101315815" "101316241" "101316870" ...
## .. ..$ GO:0001503: chr [1:25] "101319191" "101319621" "101324677" "101326016" ...
## .. ..$ GO:0001510: chr [1:56] "101315879" "101316484" "101316654" "101317321" ...
## .. ..$ GO:0001525: chr [1:67] "101315516" "101316044" "101316437" "101317073" ...
## .. ..$ GO:0001539: chr [1:51] "101316497" "101317011" "101318395" "101318705" ...
## .. ..$ GO:0001558: chr [1:37] "101316049" "101316139" "101316372" "101316688" ...
## .. ..$ GO:0001568: chr [1:74] "101315516" "101316044" "101316437" "101317073" ...
## .. ..$ GO:0001578: chr [1:58] "101315537" "101315853" "101315972" "101316059" ...
## .. ..$ GO:0001580: chr [1:11] "101317618" "101318194" "101318776" "101320041" ...
## .. ..$ GO:0001654: chr [1:46] "101316343" "101316644" "101317476" "101318045" ...
## .. ..$ GO:0001678: chr [1:11] "101322282" "101325456" "101326457" "101327365" ...
## .. ..$ GO:0001708: chr [1:27] "101317812" "101318912" "101320335" "101320674" ...
## .. ..$ GO:0001738: chr [1:4] "101326058" "101328570" "101330480" "101339398"
## .. ..$ GO:0001754: chr [1:5] "101318370" "101326924" "101330324" "101330927" ...
## .. ..$ GO:0001775: chr [1:185] "101315662" "101315938" "101316111" "101316120" ...
## .. ..$ GO:0001776: chr [1:4] "101321590" "101323139" "101329800" "101337728"
## .. ..$ GO:0001816: chr [1:107] "101315608" "101316515" "101316915" "101317221" ...
## .. ..$ GO:0001817: chr [1:98] "101315608" "101316515" "101316915" "101317221" ...
## .. ..$ GO:0001837: chr [1:3] "101323107" "101333363" "109547263"
## .. ..$ GO:0001881: chr [1:10] "101322812" "101323360" "101323714" "101334010" ...
## .. ..$ GO:0001919: chr [1:2] "101334010" "117313416"
## .. ..$ GO:0001920: chr "101334010"
## .. ..$ GO:0001932: chr [1:212] "101315937" "101316002" "101316372" "101316388" ...
## .. ..$ GO:0001933: chr [1:45] "101316388" "101316596" "101317190" "101317572" ...
## .. ..$ GO:0001934: chr [1:130] "101316002" "101316372" "101316545" "101317224" ...
## .. ..$ GO:0001941: chr [1:12] "101323758" "101324658" "101326314" "101327064" ...
## .. ..$ GO:0001944: chr [1:78] "101315516" "101316044" "101316437" "101317073" ...
## .. ..$ GO:0001993: chr [1:3] "101316390" "101326019" "101330079"
## .. ..$ GO:0002009: chr [1:23] "101316112" "101317529" "101317812" "101319445" ...
## .. ..$ GO:0002025: chr [1:3] "101316390" "101326019" "101330079"
## .. ..$ GO:0002064: chr [1:12] "101317665" "101318352" "101318526" "101321794" ...
## .. ..$ GO:0002065: chr [1:2] "101329679" "101339443"
## .. ..$ GO:0002067: chr "101339443"
## .. ..$ GO:0002088: chr [1:21] "101318045" "101318323" "101318352" "101319895" ...
## .. ..$ GO:0002093: chr [1:3] "101315722" "101335735" "117308539"
## .. ..$ GO:0002097: chr [1:18] "101316435" "101318322" "101319406" "101319432" ...
## .. ..$ GO:0002098: chr [1:17] "101316435" "101318322" "101319406" "101321637" ...
## .. ..$ GO:0002143: chr [1:6] "101319406" "101337228" "101339889" "109552667" ...
## .. ..$ GO:0002154: chr [1:3] "101322230" "101338642" "101339628"
## .. ..$ GO:0002181: chr [1:88] "101315671" "101315674" "101316145" "101316335" ...
## .. ..$ GO:0002218: chr [1:37] "101316515" "101316747" "101318035" "101318757" ...
## .. ..$ GO:0002221: chr [1:31] "101316515" "101316747" "101318035" "101318757" ...
## .. ..$ GO:0002224: chr [1:23] "101316747" "101318757" "101319483" "101319774" ...
## .. ..$ GO:0002250: chr [1:96] "101315614" "101316120" "101316872" "101317221" ...
## .. ..$ GO:0002252: chr [1:129] "101315614" "101315656" "101315662" "101316337" ...
## .. ..$ GO:0002253: chr [1:115] "101315656" "101315672" "101316146" "101316337" ...
## .. ..$ GO:0002260: chr [1:3] "101321590" "101329800" "101337728"
## .. ..$ GO:0002376: chr [1:633] "101315493" "101315608" "101315614" "101315656" ...
## .. ..$ GO:0002429: chr [1:64] "101315672" "101316146" "101316872" "101317458" ...
## .. ..$ GO:0002443: chr [1:70] "101315614" "101316374" "101318998" "101319565" ...
## .. .. [list output truncated]
## ..@ readable : logi FALSE
## ..@ termsim : num[0 , 0 ]
## ..@ method : chr(0)
## ..@ dr : list()To convert it into a “real” table, DO NOT use as.data.frame() which only returns the significant terms, use tb@result.
Also some columns in tb@result are not in the proper format, e.g. "GeneRatio" and "BgRatio" where numbers should be in numeric mode while not characters. In GSEAtraining, there is a add_more_columns() which adds more columns to the tb@result table.
tb2 = add_more_columns(tb)
head(tb2@result)## ID Description
## GO:0006188 GO:0006188 IMP biosynthetic process
## GO:0032543 GO:0032543 mitochondrial translation
## GO:0071900 GO:0071900 regulation of protein serine/threonine kinase activity
## GO:0120036 GO:0120036 plasma membrane bounded cell projection organization
## GO:0006473 GO:0006473 protein acetylation
## GO:0046040 GO:0046040 IMP metabolic process
## GeneRatio BgRatio RichFactor FoldEnrichment zScore pvalue
## GO:0006188 3/453 10/13726 0.30000000 9.090066 4.727800 0.003603571
## GO:0032543 5/453 33/13726 0.15151515 4.590943 3.815372 0.004236004
## GO:0071900 7/453 66/13726 0.10606061 3.213660 3.330261 0.005916926
## GO:0120036 26/453 473/13726 0.05496829 1.665551 2.721295 0.007733292
## GO:0006473 3/453 13/13726 0.23076923 6.992359 3.993227 0.007978294
## GO:0046040 3/453 13/13726 0.23076923 6.992359 3.993227 0.007978294
## p.adjust qvalue
## GO:0006188 0.943299 0.943299
## GO:0032543 0.943299 0.943299
## GO:0071900 0.943299 0.943299
## GO:0120036 0.943299 0.943299
## GO:0006473 0.943299 0.943299
## GO:0046040 0.943299 0.943299
## geneID
## GO:0006188 101315901/101322662/101323582
## GO:0032543 101325303/101324614/101327888/101317961/101328773
## GO:0071900 101330246/101325657/117313024/117308306/101338481/101321085/101334943
## GO:0120036 101315722/101332080/117310101/101337789/101335043/101327264/101333027/101335978/101331724/101329010/101322494/101319665/101330777/101337069/101323465/101321040/101339730/101338906/101321994/101334024/101322276/101319062/101336302/109547263/101317632/117308159
## GO:0006473 101322089/101319574/117309525
## GO:0046040 101315901/101322662/101323582
## Count n_hits n_genes gs_size n_totle log2_fold_enrichment z_score
## GO:0006188 3 3 453 10 13726 3.1842908 4.727800
## GO:0032543 5 5 453 33 13726 2.1987904 3.815372
## GO:0071900 7 7 453 66 13726 1.6842172 3.330261
## GO:0120036 26 26 453 473 13726 0.7359997 2.721295
## GO:0006473 3 3 453 13 13726 2.8057792 3.993227
## GO:0046040 3 3 453 13 13726 2.8057792 3.993227Practice
Practice 1
Use the DE gene list in webgestalt_example_gene_list.rds to perform ORA on GO/KEGG/MSigDB gene sets.
genes = readRDS(system.file("extdata", "webgestalt_example_gene_list.rds",
package = "GSEAtraining"))