Topic 4-01: CePa
Zuguang Gu z.gu@dkfz.de
2025-03-02
Source:vignettes/topic4_01_CePa.Rmd
topic4_01_CePa.RmdCePa
The CePa package has not been updated for ~10 years. I am planning to revisit it some time this year.
It will try fix different centralities:
- equal.weight
- in.degree
- out.degree
- node betweenness
- in.reach
- out.reach
dif and bk are two gene vectors, pc is in a special format.
# around 5min
res = cepa.ora.all(dif = gene.list$dif, bk = gene.list$bk, pc = PID.db$NCI)Heatmap of adjusted p-values:
plot(res, adj.method = "BH", only.sig = TRUE)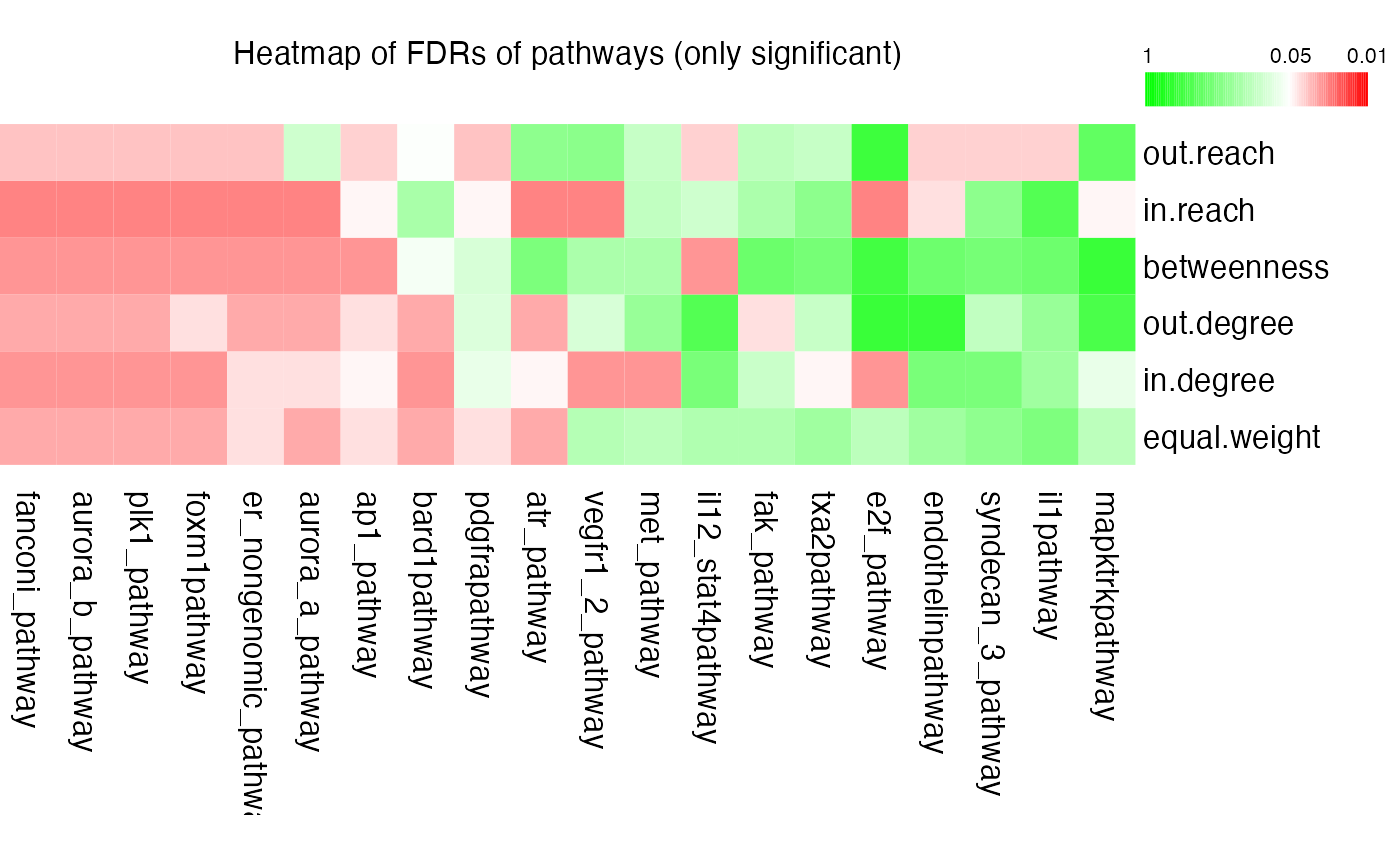
We can see “e2f_pathway” is signinicant with in-degree but not significant with out-degree. Let’s directly plot the pathway network:
Plot network for a specific pathway under a specific centrality
plot(res, "e2f_pathway", cen = "in.degree")
plot(res, "e2f_pathway", cen = "out.degree")